DO Patent: AI-Powered Tool to Convert Chemical Images into SMILES and Curate your Data
1. Overview
DO Patent is an AI tool that identifies and converts chemical images within patents and other PDF documents into SMILES strings (a convenient format for subsequent data processing).
2. Getting Started
- Account Creation/Login: Sign up for an account here or log in to your existing account.
- Select products of interest: Upon logging in, you will land on the Products page (aka the main page in Settings) where you can select which product you want to use
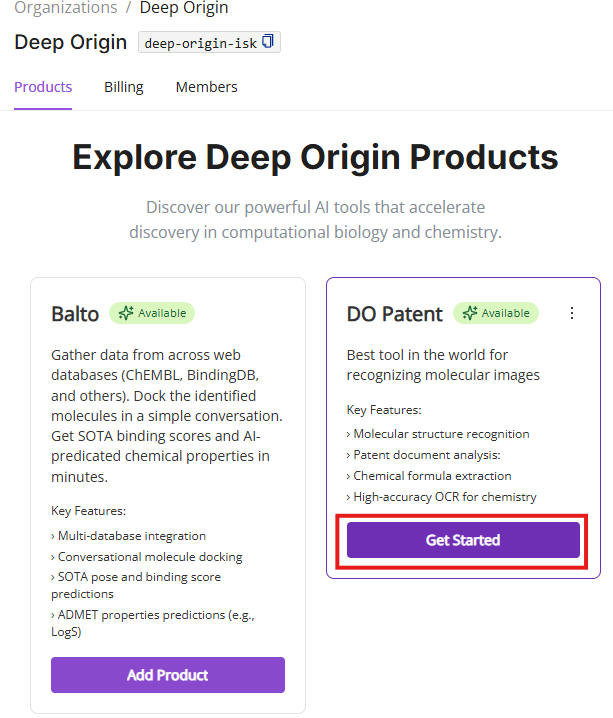
- Initial DO Patent Interface: Upon logging in, you’ll be presented with the DO Patent interface shown below, which is organized into distinct panels (described below).
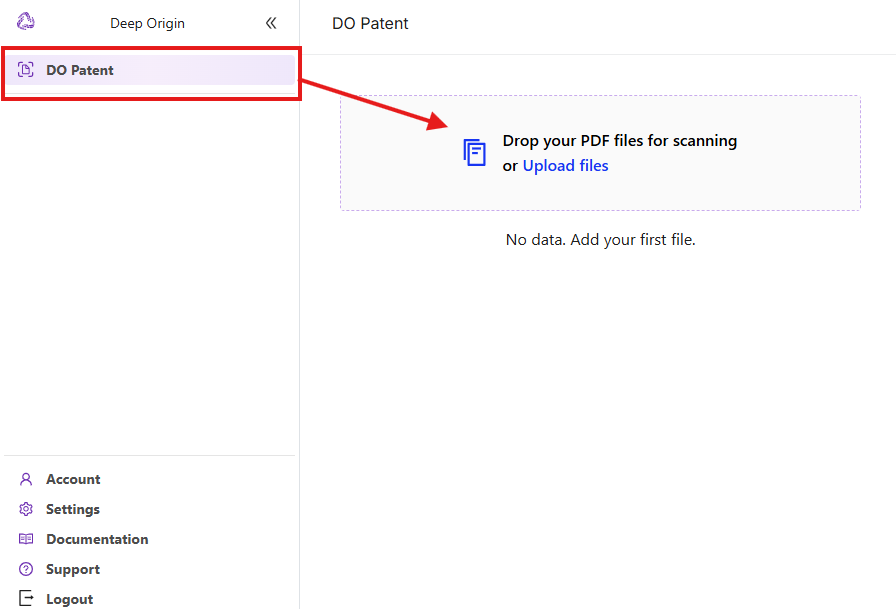
- DO Patent selection: If the interface above is not what you see, click on DO Patent in the left menu to switch to DO Patent app
3. DO Patent - User Interface and Functionality
DO Patent offers a simple solution for converting chemical images within documents of interest (patents, journal articles) into SMILES strings, a universal format for encoding small molecules. The tool also provides the original images extracted from PDF documents and confidence scores for each recognition event to make data curation straightforward.
3.1 Jobs view
Jobs view consists of two segments:
- Upload panel
- Jobs list
The upload panel allows you to input the desired documents for analysis while the Jobs list gives key identifiers to locate the desired jobs, see the jobs status, and execute necessary operations.
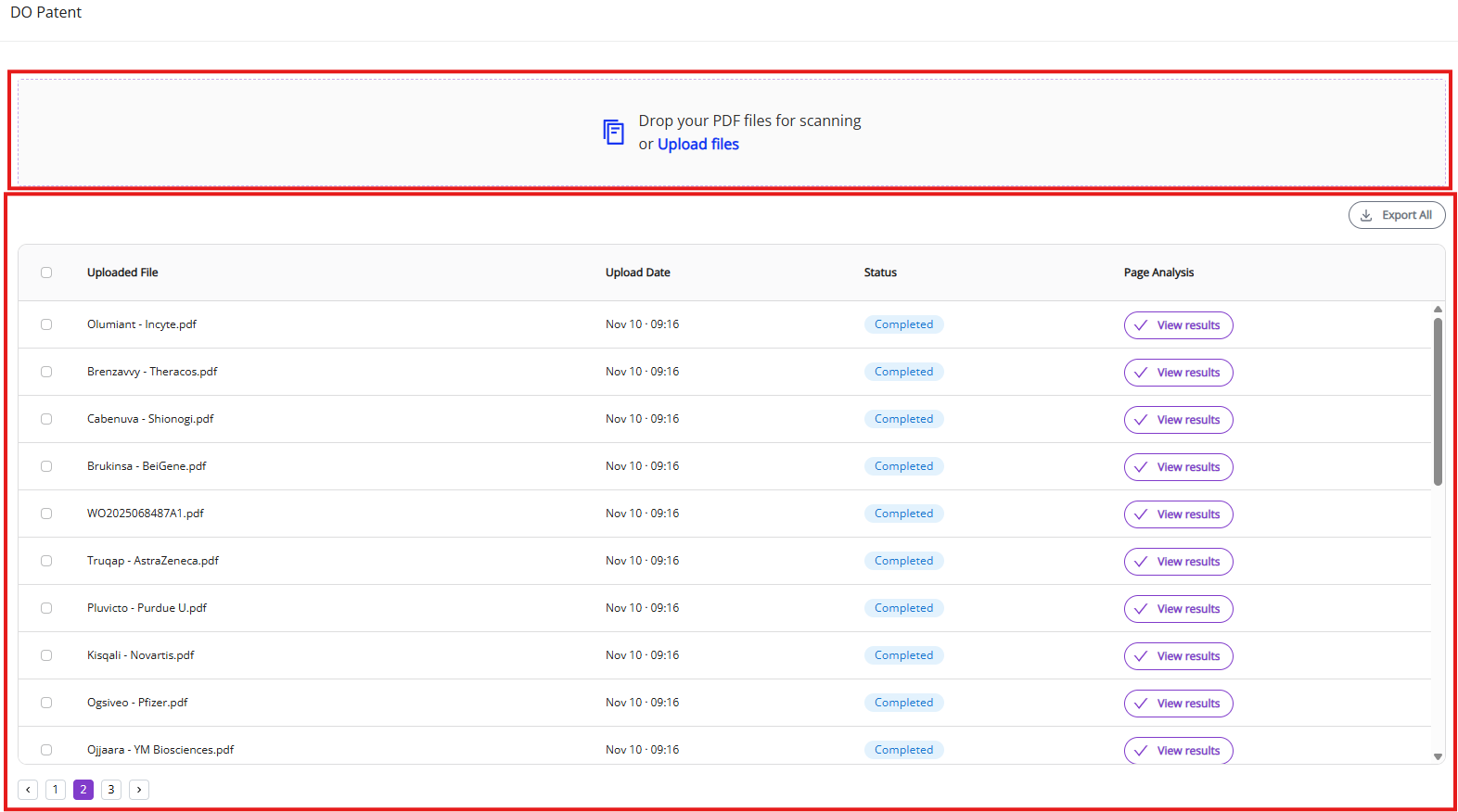
3.1.1 Upload panel and input parameters
You can upload PDF files either by dragging files into upload panel or by clicking on “Upload files” link and selecting the PDF document in the file browser.
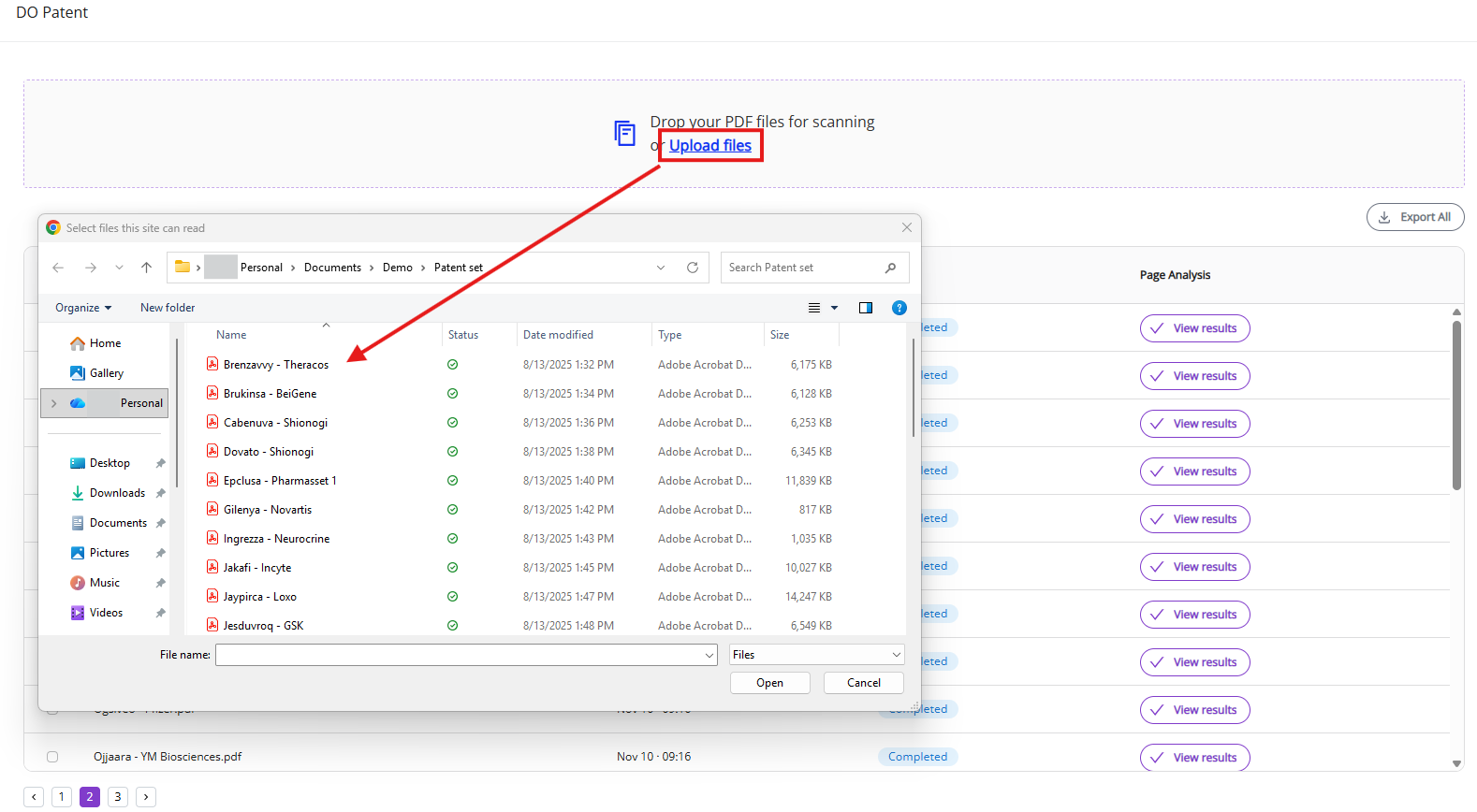
Key input parameters are outlined in the list below:
- Document type: Any document (patent or journal article) with chemical images
- Input format: PDF only
- Batch input: Supported
- Size limit: 1 Gb per document
- Color support: DO Patent supports analysis of colored chemical images. The best results are achieved with patent-like documents known for length and grainy black & white images
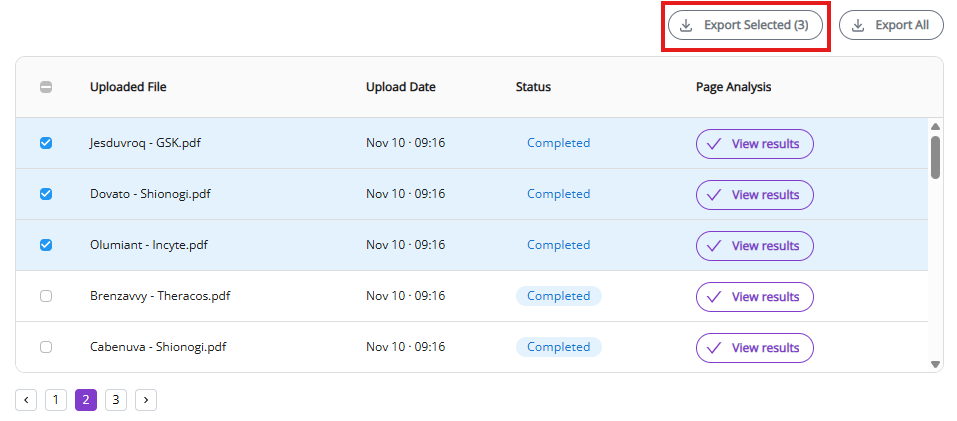
3.1.2 Jobs list
The jobs list allows you to see a list of past and active jobs and execute certain operations with active and completed processes (see below). It is sorted by default in newest to oldest order.
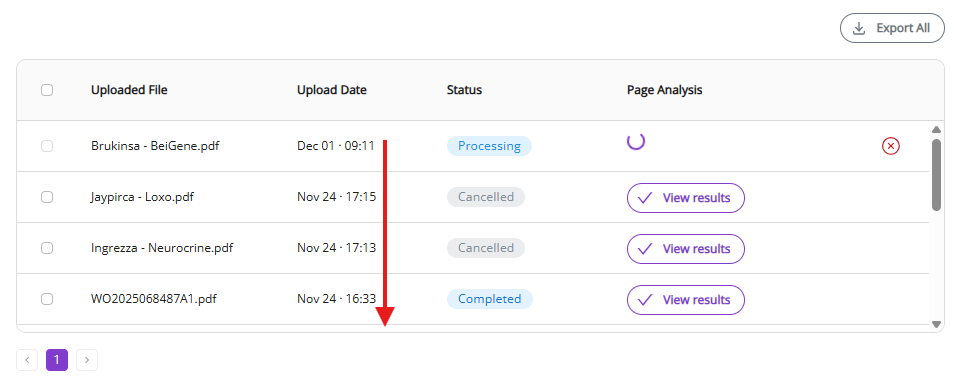
“Uploaded File” and “Upload date” allow you to quickly identify documents that you need.
“Status” and “Page Analysis” give you parameters to monitor the process (see below for more details).
“View results” button (appears as soon as document processing is complete) will open the Results View (see below) where you can view all processed molecules and edit them, if necessary.
Cancel button is shown only for active processes. It aborts the active process.
Export All and Export Selected allow you to download the processed document results (see below).
3.1.3 Execution Parameters
Key execution parameters are outlined in the list below:
- Time to completion: It takes about 30 min to process an average patent (120 pages, black & white). The actual completion time depends on document resolution, density of chemical images and presence of additional colors.
- Parallel processing: Supported. Multiple patents can be uploaded for processing at once, however total time to finish may vary.
During document analysis, DO Patent can generate the following states:
- In Queue: The file has been successfully uploaded and is waiting for analysis to start.
- Processing: The file is being analyzed by our AI engine.
- Completed: The document analysis was successful and the resulting Excel file is ready for download.
- InsufficientFunds: This status is likely caused by achieving one of two limits:
- Free pages limit
- Solution: Add a payment method
- Available credit limit
- Solution: Contact our customer support at support@deeporigin.com to increase your available credit
- Free pages limit
- Failed: Failed runs are rare. Please, contact us at support@deeporigin.com if you experience a failed job. Often errors are just flukes caused by cosmic rays that are resolved by resubmission. We do not count pages for failed runs.
- Cancelled: Document processing has been cancelled manually by the user.
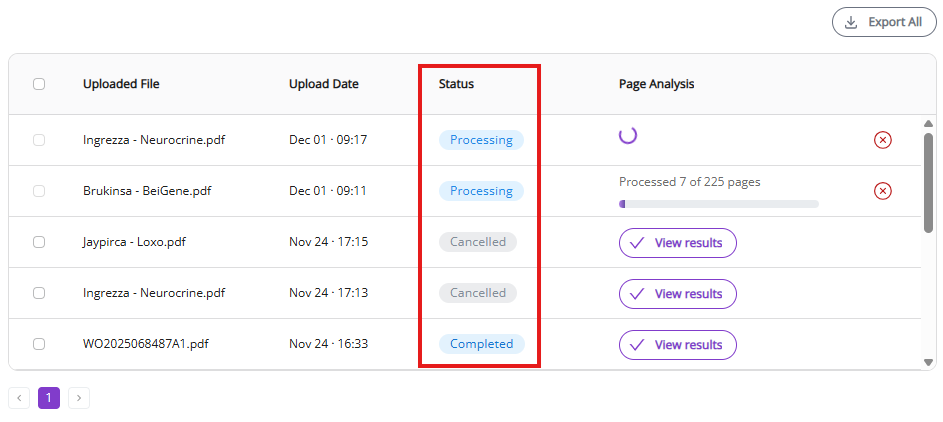
3.1.4 Job cancellation
The cancel button is shown only for active processes. It aborts the active process. Partial charges will be posted to your account according to the number of processed pages. Partial results can be exported with Export All and Export Selected buttons or can be inspected and edited by clicking on View Results.
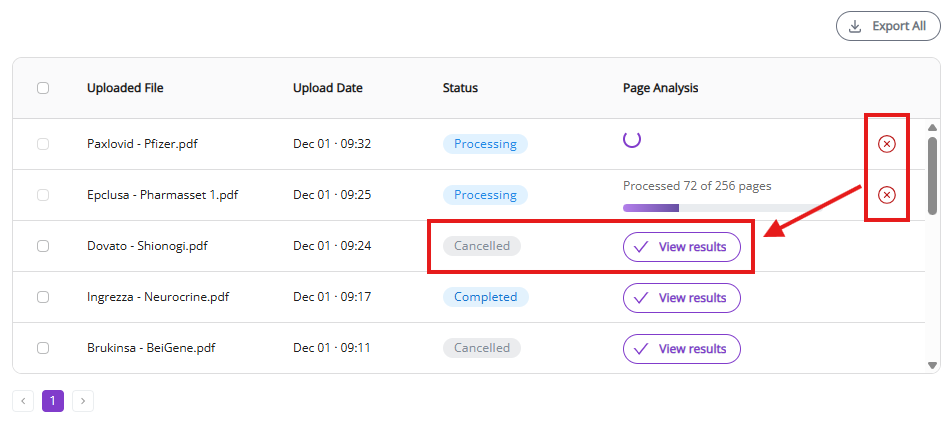
3.1.5 Export from the Jobs View
There are two options to export for the Jobs View:
- Export all: Export all jobs on the active jobs page as separate Excel files. The download will start automatically once “Export all” is clicked with one Excel file per processed document.
- Export selected: Export selected jobs as Excel files. The “Export selected” button appears once you have selected at least one job. The download of all selected jobs will start automatically once “Export selected” is clicked, with one Excel file per processed document.
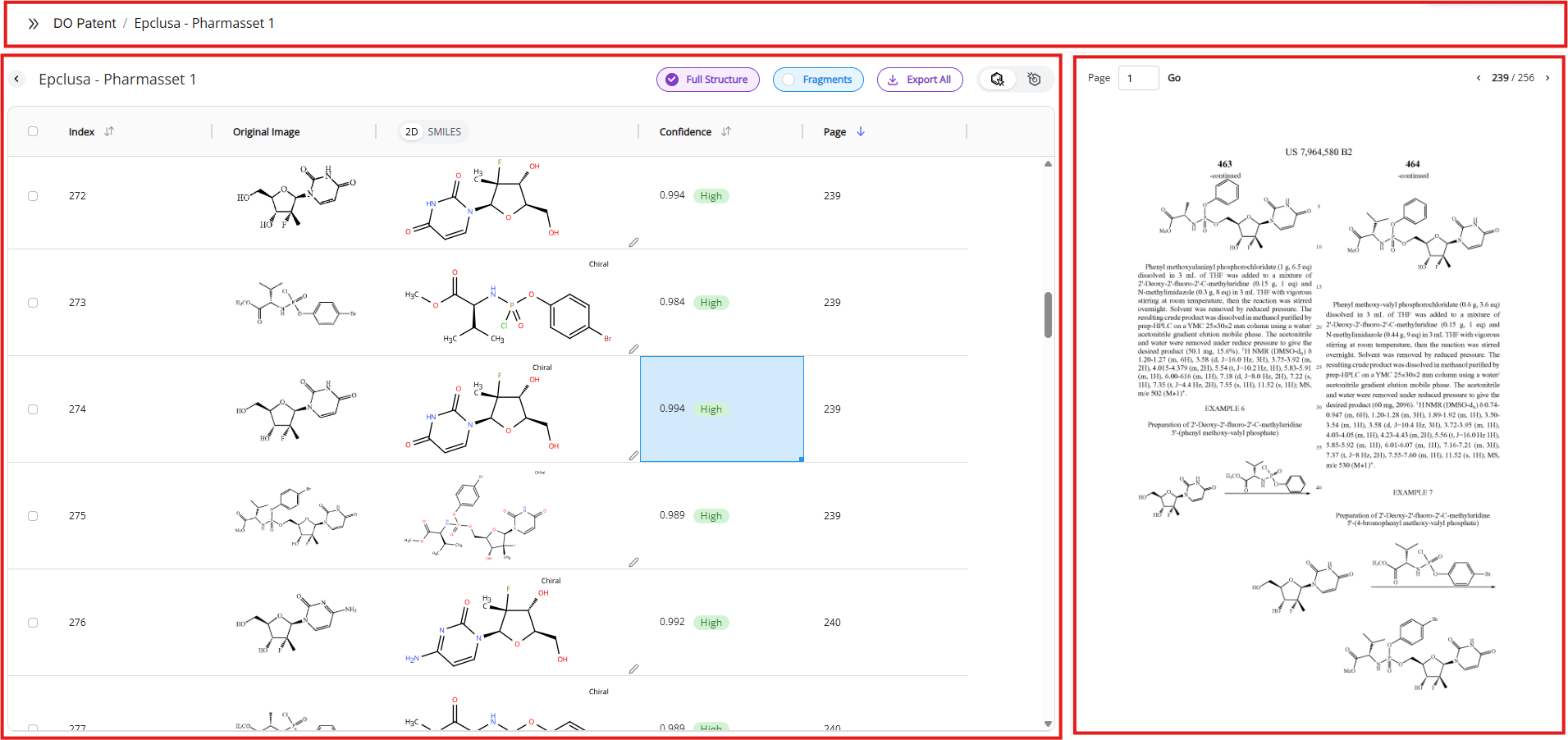
3.2 Results View
“View Results” button shows up in the Jobs view as soon as the document is successfully processed. Clicking on this button will open the Results View where you can see a full list of recognized molecules and curate your data.
The results view consist of the following segments:
- Navigation, structural modes and export
- Table
- PDF viewer
- DO Draw (see below)
3.2.1 Navigation
Basic navigation shows the name of the opened document and a chevron for returning to the Jobs view.
3.2.2 Structural modes
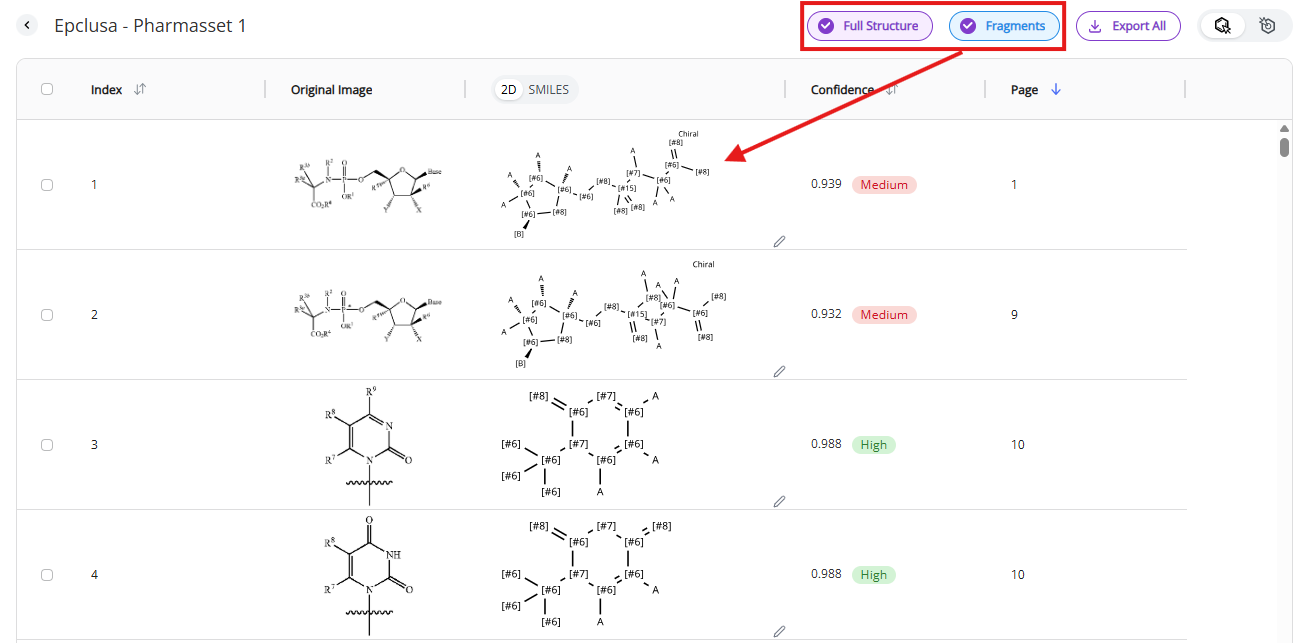
Filters control the content shown in the table or how molecules are visualized:
- Full molecules: full molecules are shown in the table when this mode is activated. Molecules without variable substituents or open valences are classified as “Full Molecules”
- Note: DO Patent shows by default only Full Molecule
- Fragments: fragments and Markush structures are shown in the table when this mode is activated. Any molecule with variable substituents or open valences are classified as “Fragments”
- Note: it is rare for our algorithm to misclassify full molecules as fragments but possible. We recommend always checking fragments with high confidence scores (>0.92) if they were misclassified.
- Kekulization toggle: fragments containing aromatic rings can be toggled between kekulized representation (circle) and non-kekulized representation (alternating single and double bonds)
- Note: if you have a mix of kekulized and non-kekulized structures in the recognized document, the kekulization operation will be applied to the non-kekulized fraction of molecules in an irreversible fashion
3.2.3 Export from the Results View
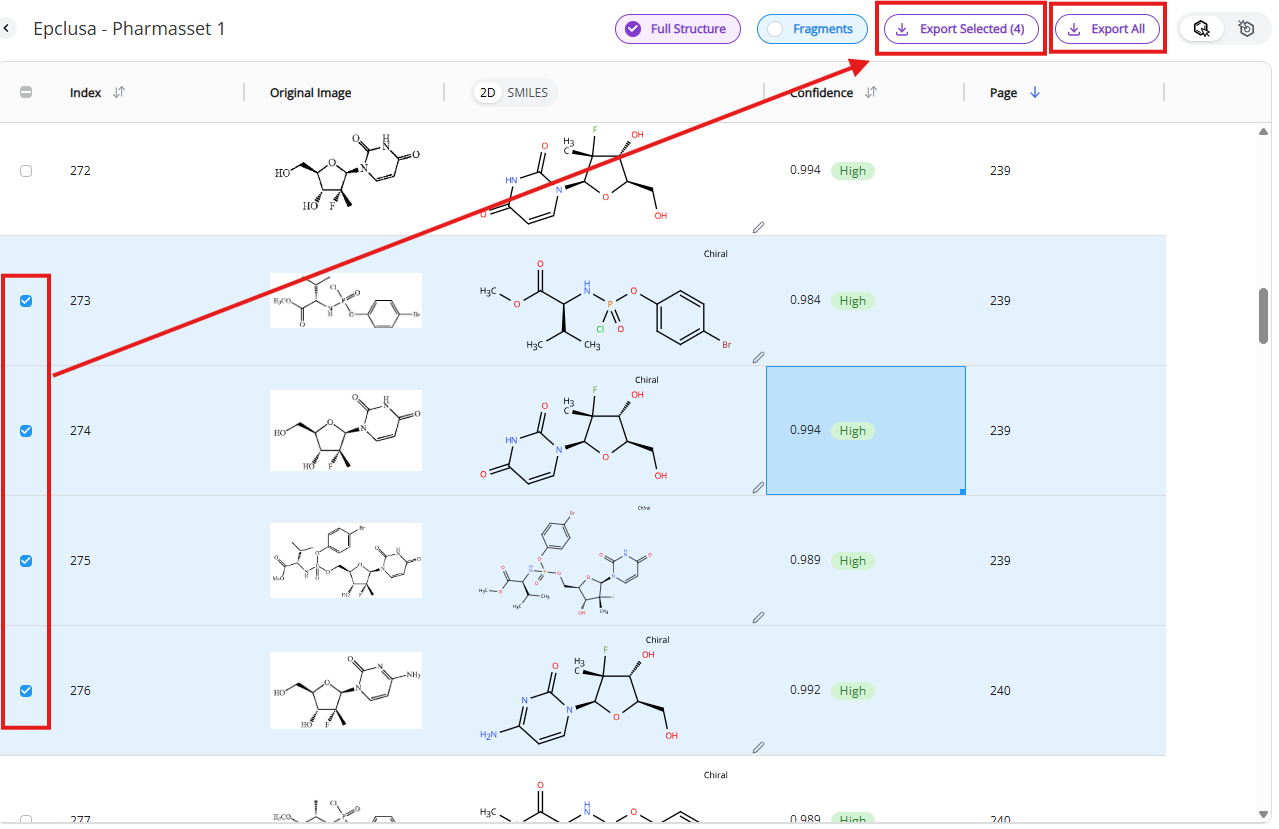
There are ways to export data from the Results view:
- Export all: Export all rows as an Excel file. The download will start automatically after “Export all” is clicked.
- Export selected: Export selected rows as an Excel file. “Export selected” button appears once you select at least one row. The download will start automatically after “Export selected” is clicked.
3.2.4 Table View
Table view allows you to curate your data and edit molecular structures.
The table view consists of the following columns:
- ID
- This column approximates the order in which molecules and fragments appear in the document
- Original image
- This column contains the image our algorithm recognized as a molecule and extracted in the document. Direct comparison between the original image and the recognized molecule facilitates data curation.
- Note: Clicking on the row will show the exact page in the PDF viewer from which the molecule was extracted so you can review broader context associated with the molecule.
- This column contains the image our algorithm recognized as a molecule and extracted in the document. Direct comparison between the original image and the recognized molecule facilitates data curation.
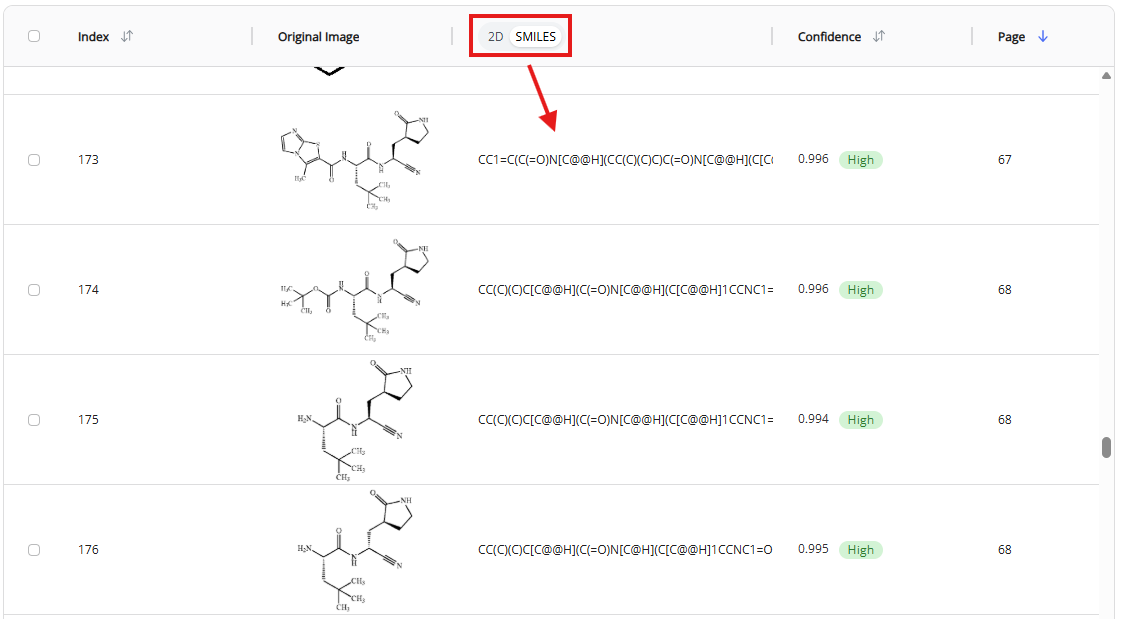
- 2D-to-SMILES toggle
- This toggle allows to switch between 2D rendering of recognized SMILES and the SMILES format for each recognized molecule
- Confidence score
- This score shows the confidence level of our AI engine for each recognized molecule. There are three confidence levels:
- High (>0.98 confidence score) - Likely accurate structure
- Note: Fraction of molecules with high confidence score depends on the document type and formatting. On average, 73% of molecules in the US patents are recognized with the high confidence score.
- Note: See the DO Patent accuracy section for more details about segmentation and recognition accuracies. Molecules with “High” confidence tag show 97.3% accuracy per molecule and 99.9% accuracy for individual structural elements (atoms and bonds). The molecule was considered inaccurate when either one atom or one bond was recognized incorrectly.
- Medium (0.92-0.98 confidence score) - Needs manual review
- Note: The fraction of molecules with medium confidence scores depends on the document type and formatting. On average, 22% of molecules in the US patents in our test set are recognized with the medium confidence score.
- Note: Recognition accuracy is highly dependent on document formatting (see 3.5.3 Recognition accuracy - Medium confidence)
- Low (<0.92 confidence score) - Consider discarding
- High (>0.98 confidence score) - Likely accurate structure
- This score shows the confidence level of our AI engine for each recognized molecule. There are three confidence levels:
- Page
- This column lists the page in the document from which its molecular structure was identified and recognized
- Note: You can also click on the row to see the exact page in the PDF viewer
By default, the table is sorted by the page number. However, you can sort the page by structure ID or confidence score.
3.2.5 PDF viewer
The PDF viewer shows the source document from which recognized molecules originated. You can navigate to a specific page by entering the page number or by clicking on the left and right chevrons.
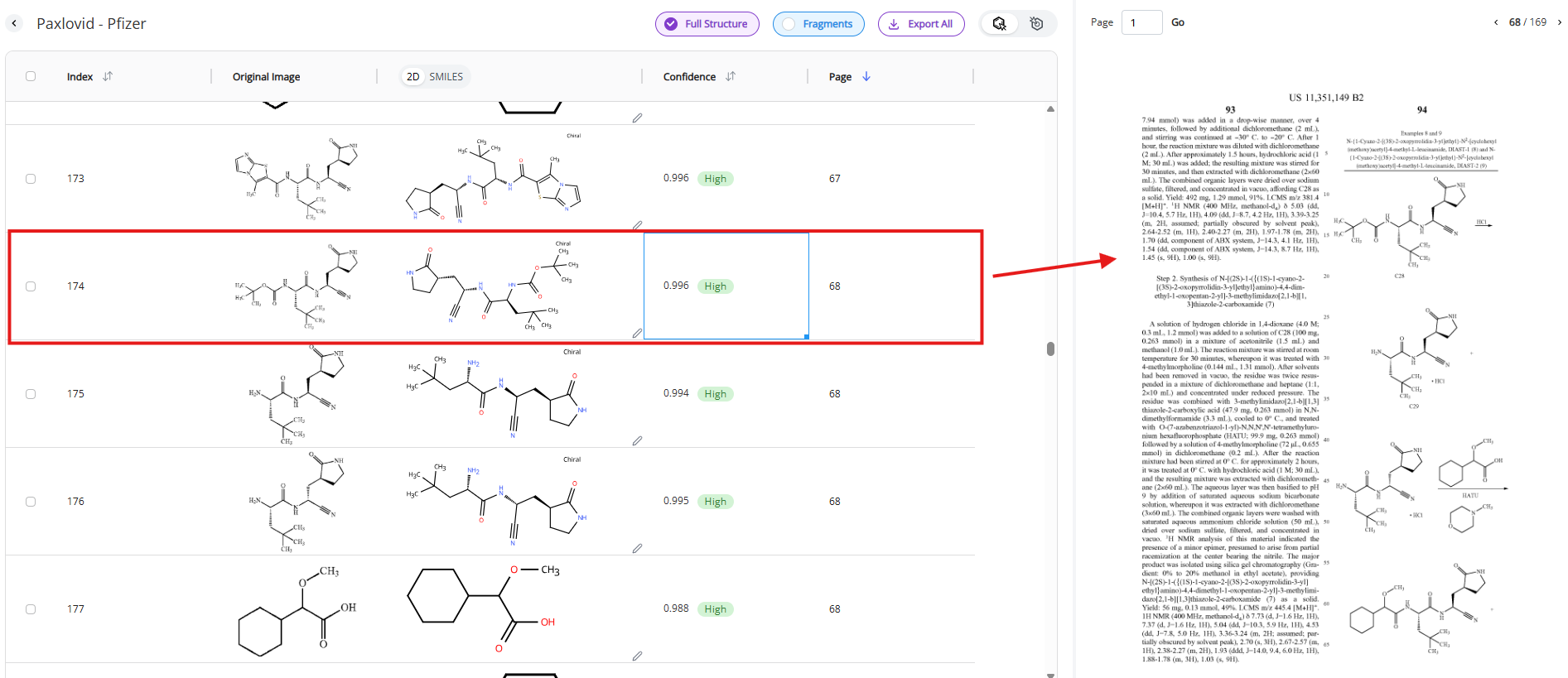
Selecting a specific row in the table navigates to the specific page in the document where the molecule in the row was found. This function is helpful if you need to review additional information related to the table.
3.3 Chemical structure and SMILES editing
DO Patent allows users to edit molecular structures and SMILES strings, if necessary.
3.3.1 Editing chemical structures with DO Draw
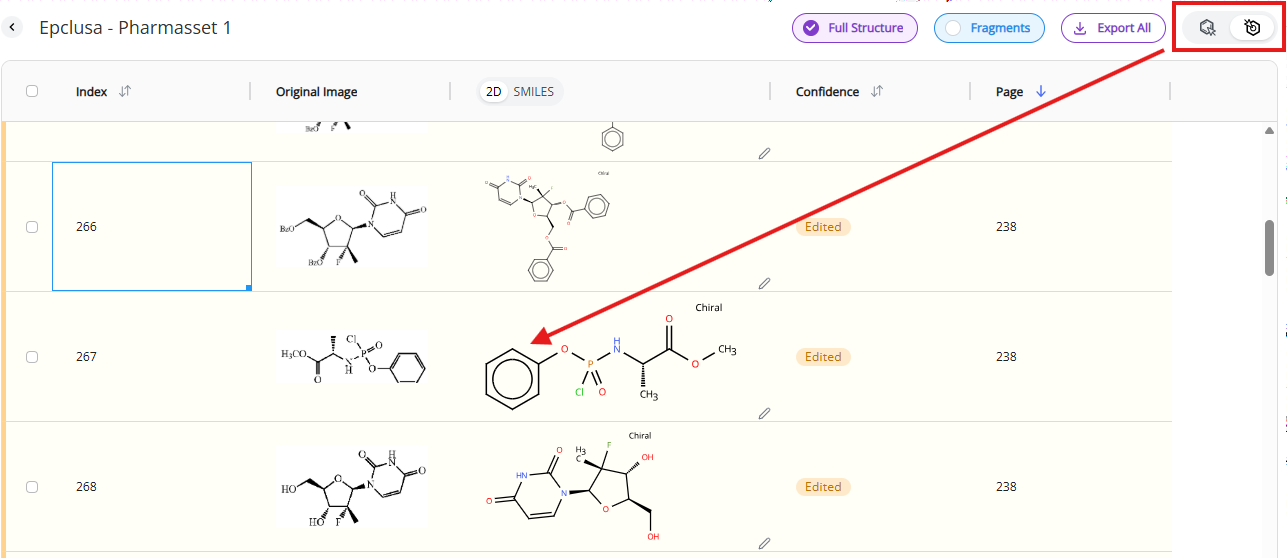
Double clicking on a 2D-rendered image of the molecule will open DO Draw, Deep Origin’s molecular editor.
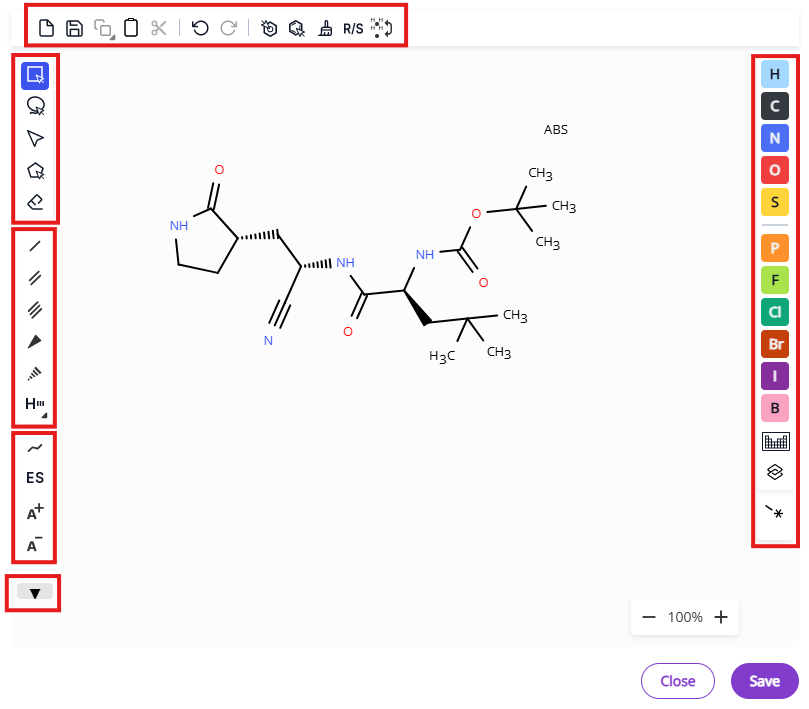
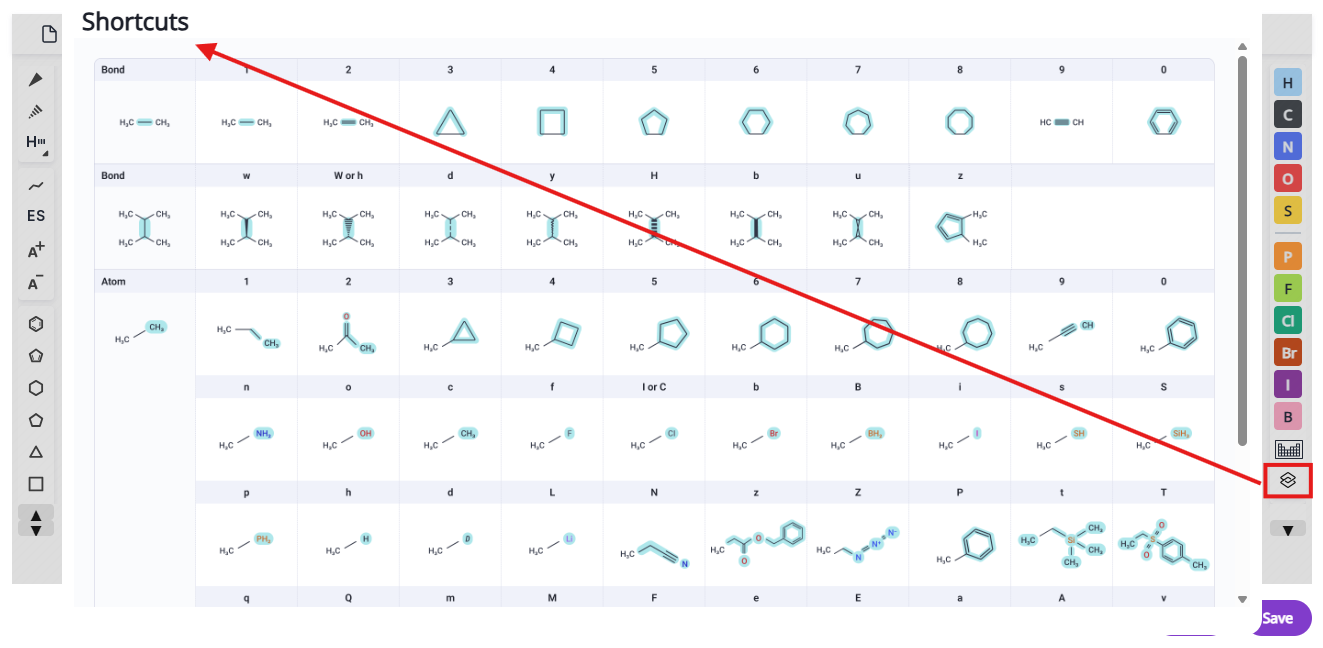
The editor contains necessary tools and several shortcuts for common moieties to facilitate editing of molecular structures. Editor tools are grouped in several menus:
- Export and bulk editing tools: Export molecule on the canvas, undo/redo operations, aromatize/dearomatize operations, clean up structure, calculate R/S designation, add/remove explicit hydrogen.
- Selection and deletion tools: Hand tool, rectangle selection, lasso selection, fragment selection, eraser.
- Bond types: Single bond, double bond, triple bond, single bond up, single bond down, hydrogen bond.
- Additional bond types are also available under the hydrogen bond expansion menu - aromatic bond, dative bond, any bond, undefined single bond, undefined double bond.
- Charges and stereochemistry: Chain tool, advanced stereochemistry, positive charge, negative charge.
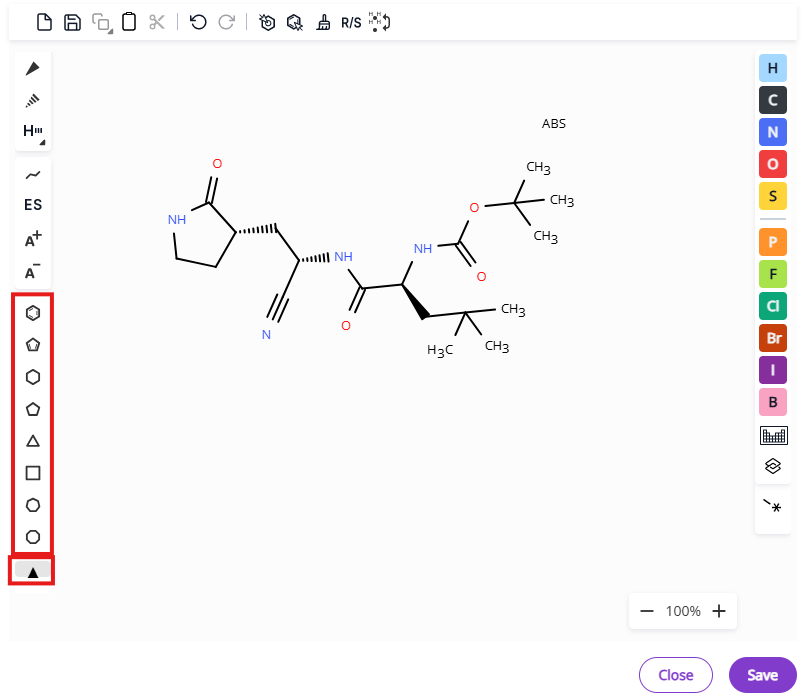
- Common rings: Benzene, cyclopentadiene and three- to eight-membered aliphatic rings.
- Atom types: Common atoms and periodic table
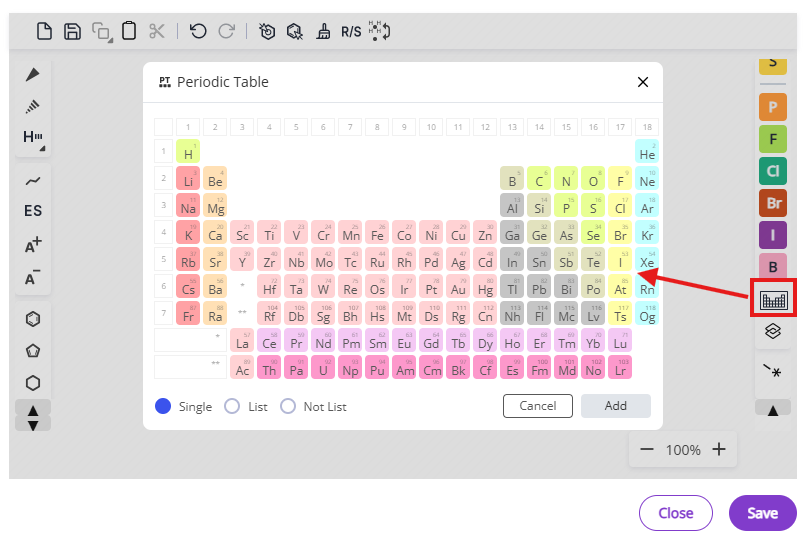
List of available shortcuts is accessible via the shortcuts button and also below:
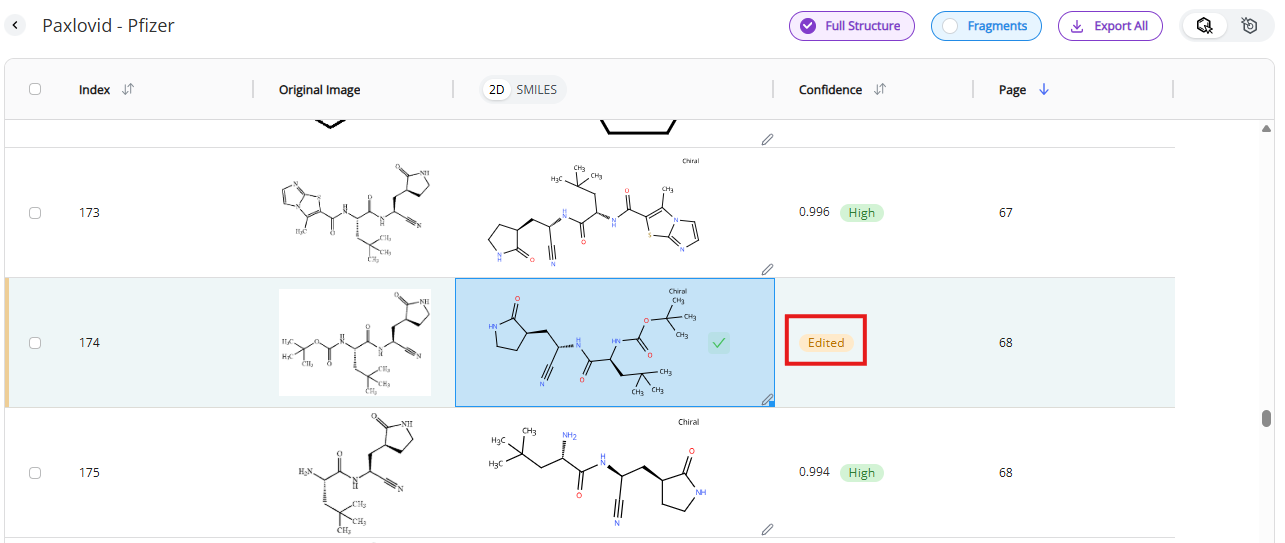
Clicking on the “Save” button will update the record. This is an irreversible change. Edited molecules will receive “edited” tag in the confidence column that will replace the original confidence score
Clicking on “Cancel” button will discard any changes made to the molecule.
3.3.2 Editing SMILES strings
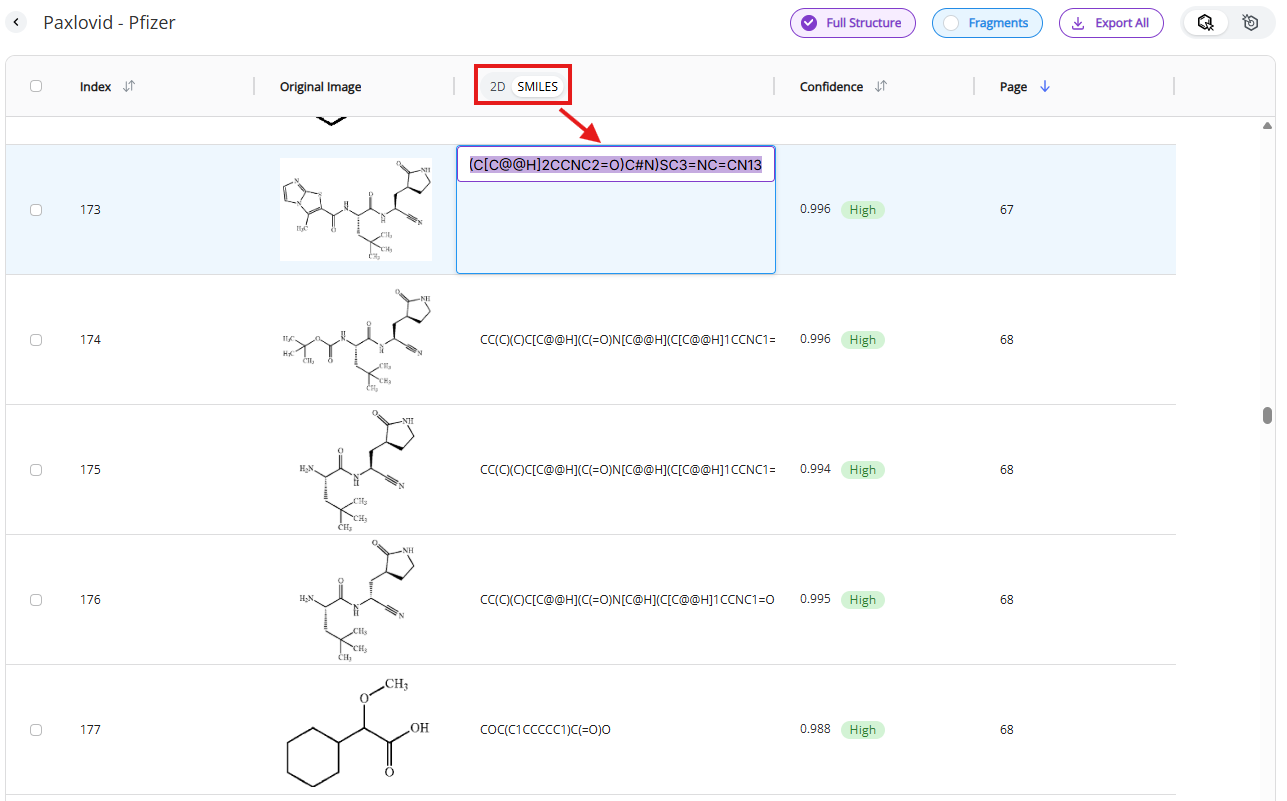
Switching from 2D rendered molecules to SMILES strings will visualize SMILES strings. Double clicking on the SMILES string will open text editor within the cell.
3.4 Export format
Processed data can be extracted from either the Jobs view (see section 3.1.5 Export from the Jobs view) or the Results view (see section 3.2.3 Export from the Results view). The data is exported in the .xlsx format (Excel). The size of file can reach 100 Mb and depends on the number of chemical images in the processed PDF document.
The output format is optimized for quick data curation and subsequent import into external databases and software solutions.
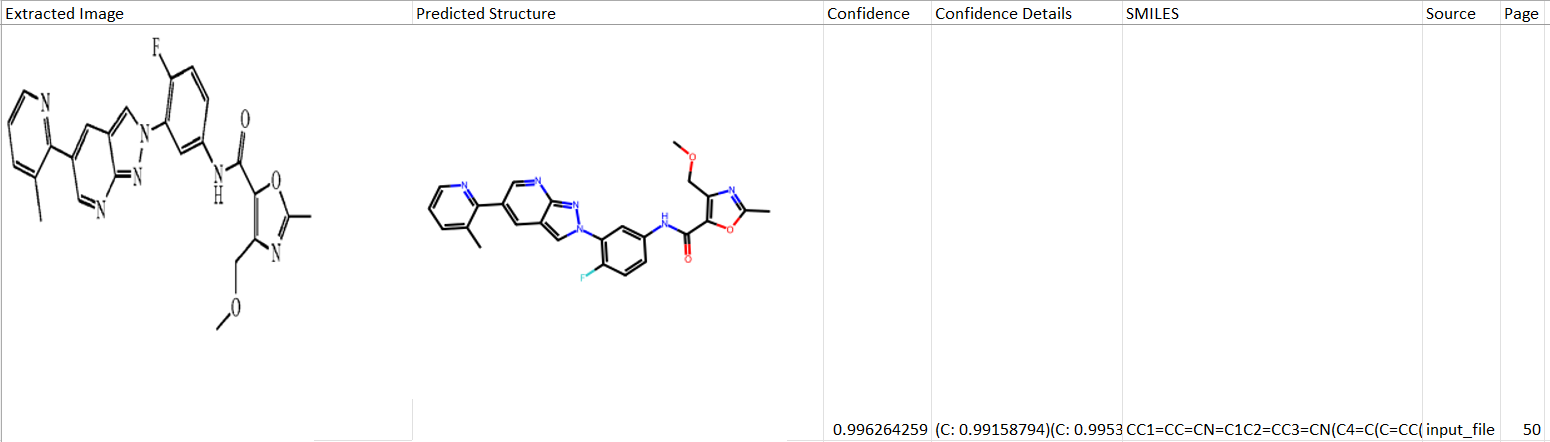
The resulting .xlsx file has the following columns:
- Structure ID: The order in which this molecule appears in the document
- Extracted Image: The original image in the PDF document as the algorithm recognized it.
- Predicted structure: 2D rendering of a chemical structure encoded in a SMILES string (see below).
- Confidence: Confidence score indicating accuracy of recognition and the need for manual data review. We recommend sorting results by the confidence score. See details below how confidence score is calculated.
- >0.98 confidence score: high likelihood of accurate recognition
- 0.92-0.98 confidence score: manual review is needed
- <0.92 confidence score: poor recognition, consider discarding result
- Confidence details: Specific recognition tokens forming the confidence score from the elements of the molecular structure.
- SMILES: 1D representation of the molecule predicted by the algorithm. This is a standard format for data import across all scientific software solutions.
- Source: Name of the original PDF document.
- Page: Page number of the recognized image of the molecule.
3.5 DO Patent accuracy
DO patent consists of two systems: segmentation and recognition. Segmentation module identifies and classifies images that contain molecules. Recognition module looks at the image trying to predict SMILES string that would fit the image.
This accuracy analysis was conducted by an experienced medicinal chemist manually looking at each page of a PDF document and comparing it to the segmentation and recognition results. 25 random US patents were selected for this exercise to capture diversity of formatting styles. Criteria for selection were type of patent, document size, filing company, market share of a drug and therapeutic modality.
3.5.1 Segmentation accuracy
During the segmentation accuracy analysis, extracted non-chemical images and images containing more than one chemical entity were considered “false positives”. Chemical images present in the PDF document but missing in the Results table were considered “false negatives”.
| patent ID | entity | company | number of pages | number of structures | number of false positive | segmentation accuracy, % | number of false negative | segmentation accuracy, % |
|---|---|---|---|---|---|---|---|---|
| US7838499 B2 | Brenzavvy | Theracos | 74 | 330 | 3 | 99.1% | 0 | 100.0% |
| US2022/0324863 A1 | Clinical candidate for Leishmaniasis | Novartis | 135 | 742 | 12 | 98.4% | 6 | 99.2% |
| US9447106 B2 | Brukinsa | BeiGene | 225 | 815 | 8 | 99.0% | 0 | 100.0% |
| US8410103 B2 | Cabenuva | Shionogi | 94 | 411 | 7 | 98.3% | 3 | 99.3% |
| US8039627 B2 | Ingrezza | Neurocrine | 18 | 25 | 6 | 76.0% | 0 | 100.0% |
| US9592208 B2 | Gilenya | Novartis | 9 | 2 | 0 | 100.0% | 0 | 100.0% |
| US8324208 B2 | Jesduvroq | GSK | 65 | 164 | 1 | 99.4% | 1 | 99.4% |
| US8324225 B2 | Kisqali | Novartis | 131 | 706 | 10 | 98.6% | 0 | 100.0% |
| US11351149 B2 | Paxlovid | Pfizer | 169 | 554 | 19 | 96.6% | 0 | 100.0% |
| US8129385 B2 | Dovato | Shionogi | 92 | 414 | 7 | 98.3% | 0 | 100.0% |
| US7964580 B2 | Epclusa | Pharmasset | 256 | 342 | 1 | 99.7% | 1 | 99.7% |
| US7598257 B2 | Jakafi | Incyte | 190 | 971 | 3 | 99.7% | 12 | 98.8% |
| US10342780 B2 | Jaypirca | Loxo | 179 | 872 | 6 | 99.3% | 0 | 100.0% |
| US8207125 B2 | Kyprolis | Onyx | 38 | 112 | 1 | 99.1% | 0 | 100.0% |
| US9617258 B2 | Litfulo | Pfizer | 142 | 444 | 5 | 98.9% | 0 | 100.0% |
| US8937150 B2 | Mavyret | AbbVie | 323 | 1993 | 51 | 97.4% | 0 | 100.0% |
| US7390791 B2 | Odefsey | Gilead | 29 | 91 | 4 | 95.6% | 0 | 100.0% |
| US7342118 B2 | Ogsiveo | Pfizer | 47 | 62 | 3 | 95.2% | 1 | 98.4% |
| US8486941 B2 | Ojjaara | YM Biosciences | 65 | 104 | 2 | 98.1% | 0 | 100.0% |
| US8158616 B2 | Olumiant | Incyte | 79 | 223 | 2 | 99.1% | 2 | 99.1% |
| US7427638 B2 | Otezla | Amgen | 24 | 14 | 7 | 50.0% | 0 | 100.0% |
| US10406240 B2 | Pluvicto | Purdue U | 79 | 207 | 7 | 96.6% | 6 | 97.1% |
| US8101623 B2 | Truqap | AstraZeneca | 83 | 233 | 4 | 98.3% | 0 | 100.0% |
| US8754096 B2 | Ubrelvy | Merck | 34 | 115 | 0 | 100.0% | 0 | 100.0% |
| US9309245 B2 | Xacduro | Entasis | 107 | 432 | 2 | 99.5% | 0 | 100.0% |
| Total | 2687 | 10378 | 171 | 98.4% | 32 | 99.7% |
3.5.2 Recognition accuracy - High confidence (>0.98 score)
Recognition accuracy was assessed only for full molecules (molecules without open valences or variable ligands). A molecule with a single error in a bond or an atom was considered an recognition error. Recognition of individual elements (atoms and bonds) were estimated from number of high confidence molecules and number of recognition errors. The vast majority of molecules with high confidence scores carried a single individual element that was recognized with an error.
| patent ID | entity | company | number of pages | number of full molecules | number of high confidence molecules | fraction of high confidence molecules, % | number of errors of high confidence molecules | recognition accuracy, % |
|---|---|---|---|---|---|---|---|---|
| US7838499 B2 | Brenzavvy | Theracos | 74 | 292 | 233 | 79.8% | 6 | 97.4% |
| US2022/0324863 A1 | Clinical candidate for Leishmaniasis | Novartis | 135 | 526 | 484 | 92.0% | 1 | 99.8% |
| US9447106 B2 | Brukinsa | BeiGene | 225 | 732 | 690 | 94.3% | 2 | 99.7% |
| US8410103 B2 | Cabenuva | Shionogi | 94 | 260 | 157 | 60.4% | 21 | 86.6% |
| US8039627 B2 | Ingrezza | Neurocrine | 18 | 9 | 5 | 55.6% | 0 | 100.0% |
| US9592208 B2 | Gilenya | Novartis | 9 | 2 | 2 | 100.0% | 0 | 100.0% |
| US8324208 B2 | Jesduvroq | GSK | 65 | 133 | 131 | 98.5% | 2 | 98.5% |
| US8324225 B2 | Kisqali | Novartis | 131 | 604 | 590 | 97.7% | 1 | 99.8% |
| US11351149 B2 | Paxlovid | Pfizer | 169 | 497 | 360 | 72.4% | 36 | 90.0% |
| US8129385 B2 | Dovato | Shionogi | 92 | 254 | 144 | 56.7% | 14 | 90.3% |
| US7964580 B2 | Epclusa | Pharmasset | 256 | 68 | 57 | 83.8% | 7 | 87.7% |
| US7598257 B2 | Jakafi | Incyte | 190 | 221 | 161 | 72.9% | 2 | 98.8% |
| US10342780 B2 | Jaypirca | Loxo | 179 | 548 | 458 | 83.6% | 3 | 99.3% |
| US8207125 B2 | Kyprolis | Onyx | 38 | 93 | 6 | 6.5% | 2 | 66.7% |
| US9617258 B2 | Litfulo | Pfizer | 142 | 383 | 264 | 68.9% | 1 | 99.6% |
| US8937150 B2 | Mavyret | AbbVie | 323 | 546 | 227 | 41.6% | 8 | 96.5% |
| US7390791 B2 | Odefsey | Gilead | 29 | 50 | 27 | 54.0% | 1 | 96.3% |
| US7342118 B2 | Ogsiveo | Pfizer | 47 | 1 | — | — | — | — |
| US8486941 B2 | Ojjaara | YM Biosciences | 65 | 95 | 3 | 3.2% | 0 | 100.0% |
| US8158616 B2 | Olumiant | Incyte | 79 | 123 | 89 | 72.4% | 1 | 98.9% |
| US7427638 B2 | Otezla | Amgen | 24 | 7 | 3 | 42.9% | 0 | 100.0% |
| US10406240 B2 | Pluvicto | Purdue U | 79 | 86 | 23 | 26.7% | 3 | 87.0% |
| US8101623 B2 | Truqap | AstraZeneca | 83 | 196 | 192 | 98.0% | 3 | 98.4% |
| US8754096 B2 | Ubrelvy | Merck | 34 | 61 | 52 | 85.2% | 0 | 100.0% |
| US9309245 B2 | Xacduro | Entasis | 107 | 369 | 124 | 33.6% | 5 | 96.0% |
| Total | 2687 | 6156 | 4482 | 72.8% | 119 | 97.3% | ||
| Number of atoms | 231429 | 119 | 99.95% | |||||
| Number of bonds | 185143 | 119 | 99.94% |
3.5.3 Recognition accuracy - Medium confidence (0.92-0.98 score)
The methodology for accuracy assessment of molecules with medium confidence score was similar to molecules with high confidence scores. Recognition accuracy of individual elements was not calculated because probability of molecules carrying more than one recognition error of individual elements was non-negligible.
| patent ID | entity | company | number of pages | number of full molecules | number of medium confidence molecules | fraction of medium confidence molecules, % | number of errors of medium confidence molecules | recognition accuracy, % |
|---|---|---|---|---|---|---|---|---|
| US7838499 B2 | Brenzavvy | Theracos | 74 | 292 | 41 | 14.0% | 15 | 63.4% |
| US2022/0324863 A1 | Clinical candidate for Leishmaniasis | Novartis | 135 | 526 | 26 | 4.9% | 9 | 65.4% |
| US9447106 B2 | Brukinsa | BeiGene | 225 | 732 | 40 | 5.5% | 12 | 70.0% |
| US8410103 B2 | Cabenuva | Shionogi | 94 | 260 | 84 | 32.3% | 31 | 63.1% |
| US8039627 B2 | Ingrezza | Neurocrine | 18 | 9 | 4 | 44.4% | 0 | 100.0% |
| US9592208 B2 | Gilenya | Novartis | 9 | 2 | — | — | — | — |
| US8324208 B2 | Jesduvroq | GSK | 65 | 133 | 2 | 1.5% | 1 | 50.0% |
| US8324225 B2 | Kisqali | Novartis | 131 | 604 | 14 | 2.3% | 5 | 64.3% |
| US11351149 B2 | Paxlovid | Pfizer | 169 | 497 | 74 | 14.9% | 10 | 86.5% |
| US8129385 B2 | Dovato | Shionogi | 92 | 254 | 97 | 38.2% | 34 | 64.9% |
| US7964580 B2 | Epclusa | Pharmasset | 256 | 68 | 7 | 10.3% | 1 | 85.7% |
| US7598257 B2 | Jakafi | Incyte | 190 | 221 | 47 | 21.3% | 14 | 70.2% |
| US10342780 B2 | Jaypirca | Loxo | 179 | 548 | 76 | 13.9% | 33 | 56.6% |
| US8207125 B2 | Kyprolis | Onyx | 38 | 93 | 82 | 88.2% | 56 | 31.7% |
| US9617258 B2 | Litfulo | Pfizer | 142 | 383 | 109 | 28.5% | 27 | 75.2% |
| US8937150 B2 | Mavyret | AbbVie | 323 | 546 | 311 | 57.0% | 75 | 75.9% |
| US7390791 B2 | Odefsey | Gilead | 29 | 50 | 18 | 36.0% | 4 | 77.8% |
| US7342118 B2 | Ogsiveo | Pfizer | 47 | 1 | 1 | 100.0% | 0 | 100.0% |
| US8486941 B2 | Ojjaara | YM Biosciences | 65 | 95 | 18 | 18.9% | 14 | 22.2% |
| US8158616 B2 | Olumiant | Incyte | 79 | 123 | 31 | 25.2% | 6 | 80.6% |
| US7427638 B2 | Otezla | Amgen | 24 | 7 | 4 | 57.1% | 0 | 100.0% |
| US10406240 B2 | Pluvicto | Purdue U | 79 | 86 | 48 | 55.8% | 18 | 62.5% |
| US8101623 B2 | Truqap | AstraZeneca | 83 | 196 | 3 | 1.5% | 3 | 0.0% |
| US8754096 B2 | Ubrelvy | Merck | 34 | 61 | 8 | 13.1% | 2 | 75.0% |
| US9309245 B2 | Xacduro | Entasis | 107 | 369 | 216 | 58.5% | 105 | 51.4% |
| Total | 2687 | 6156 | 1361 | 22.1% | 475 | 65.1% |
3.5.4 Recognition accuracy - Low confidence (<0.92 score)
The methodology for accuracy assessment of molecules with low confidence score was similar to molecules with high confidence scores. Recognition accuracy of individual elements was not calculated because probability of molecules carrying more than one recognition error of individual elements was non-negligible.
| patent ID | entity | company | number of pages | number of full molecules | number of low confidence molecules | fraction of low confidence molecules, % | number of errors of low confidence molecules | recognition accuracy, % |
|---|---|---|---|---|---|---|---|---|
| US7838499 B2 | Brenzavvy | Theracos | 74 | 292 | 18 | 6.2% | 10 | 44.4% |
| US2022/0324863 A1 | Clinical candidate for Leishmaniasis | Novartis | 135 | 526 | 16 | 3.0% | 12 | 25.0% |
| US9447106 B2 | Brukinsa | BeiGene | 225 | 732 | 3 | 0.4% | 3 | 0.0% |
| US8410103 B2 | Cabenuva | Shionogi | 94 | 260 | 14 | 5.4% | 5 | 64.3% |
| US8039627 B2 | Ingrezza | Neurocrine | 18 | 9 | — | — | — | — |
| US9592208 B2 | Gilenya | Novartis | 9 | 2 | — | — | — | — |
| US8324208 B2 | Jesduvroq | GSK | 65 | 133 | — | — | — | — |
| US8324225 B2 | Kisqali | Novartis | 131 | 604 | — | — | — | — |
| US11351149 B2 | Paxlovid | Pfizer | 169 | 497 | 62 | 12.5% | 26 | 58.1% |
| US8129385 B2 | Dovato | Shionogi | 92 | 254 | 13 | 5.1% | 7 | 46.2% |
| US7964580 B2 | Epclusa | Pharmasset | 256 | 68 | 4 | 5.9% | 0 | 100.0% |
| US7598257 B2 | Jakafi | Incyte | 190 | 221 | 12 | 5.4% | 8 | 33.3% |
| US10342780 B2 | Jaypirca | Loxo | 179 | 548 | 14 | 2.6% | 3 | 78.6% |
| US8207125 B2 | Kyprolis | Onyx | 38 | 93 | 6 | 6.5% | 3 | 50.0% |
| US9617258 B2 | Litfulo | Pfizer | 142 | 383 | 10 | 2.6% | 4 | 60.0% |
| US8937150 B2 | Mavyret | AbbVie | 323 | 546 | 6 | 1.1% | 6 | 0.0% |
| US7390791 B2 | Odefsey | Gilead | 29 | 50 | 5 | 10.0% | 3 | 40.0% |
| US7342118 B2 | Ogsiveo | Pfizer | 47 | 1 | — | — | — | — |
| US8486941 B2 | Ojjaara | YM Biosciences | 65 | 95 | 74 | 77.9% | 73 | 1.4% |
| US8158616 B2 | Olumiant | Incyte | 79 | 123 | 3 | 2.4% | 1 | 66.7% |
| US7427638 B2 | Otezla | Amgen | 24 | 7 | — | — | — | — |
| US10406240 B2 | Pluvicto | Purdue U | 79 | 86 | 15 | 17.4% | 11 | 26.7% |
| US8101623 B2 | Truqap | AstraZeneca | 83 | 196 | 1 | 0.5% | 1 | 0.0% |
| US8754096 B2 | Ubrelvy | Merck | 34 | 61 | 1 | 1.6% | 1 | 0.0% |
| US9309245 B2 | Xacduro | Entasis | 107 | 369 | 29 | 7.9% | 27 | 6.9% |
| Total | 2687 | 6156 | 306 | 5.0% | 204 | 33.3% |
4. Deep Origin’s User Portal
The portal interface is designed to host multiple applications (e.g., Balto - the first AI assistant for drug discovery). It is divided into two main panels (see detailed descriptions in the following sessions):
- Products and Settings panel
- Application panel
4.1 Products and Account Settings Panel
The left panel provides access to various Deep Origin products and account settings. It is divided into two segments:
Products and Settings
- Top Segment: Displays a list of your active Deep Origin products that you have activated on the Product selection page
- Bottom Segment: Provides links to:
- Account: Manage your account information (first name, last name, title, company, password). Clicking “Account” takes you to the account settings page.
- Settings: Access Deep Origin’s product selection, pricing & billing details, manage team members. (See Settings Menu for details.)
- Documentation: Direct access to this documentation
- Support: Send support email to our customer support team at support@deeporigin.com
- Logout: Log out of your Deep Origin account
You can collapse or expand this panel by clicking the double arrow (<<) next to your name.
5. Pricing
Subscription and pricing model
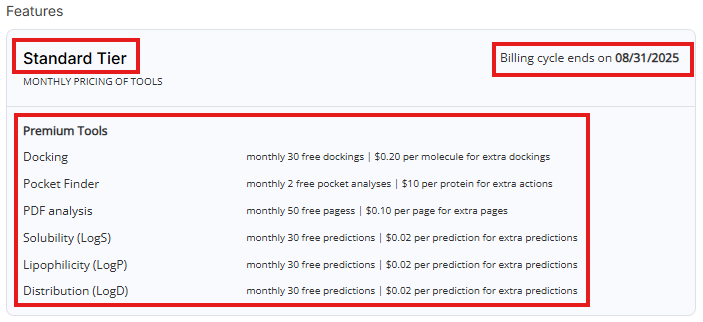
DO Patent uses pay-per-use pricing. Creating an account, monthly subscription and the analysis of the first 50 pages each month are FREE. Pages exceeding monthly free page limit will be processed according to your Pricing Tier (see below):
- Standard Tier: $0.10 per page
- Academic Tier: $0.06 per page
Free Page Count
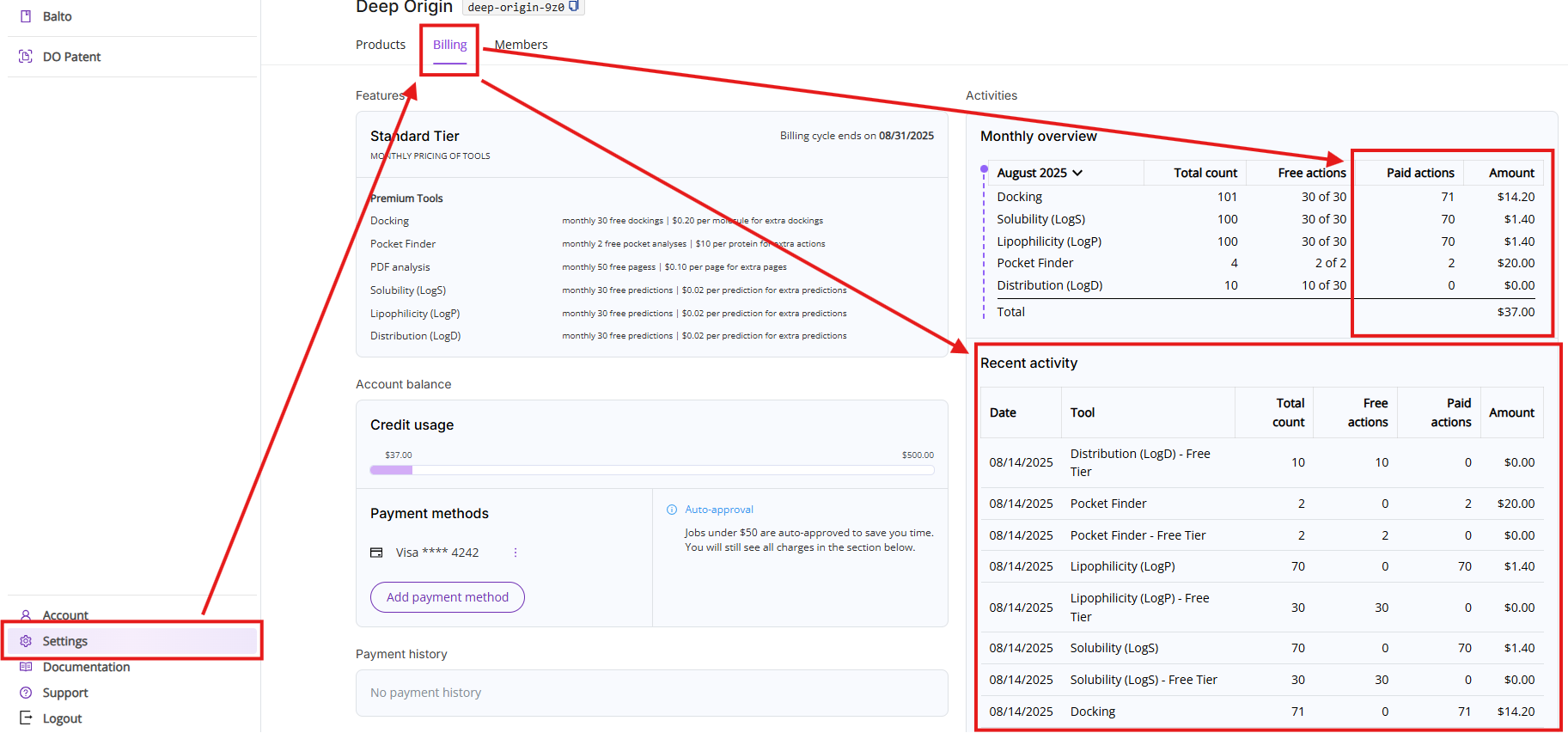
You can access remaining free pages by clicking on Settings and then on the Billing tab.
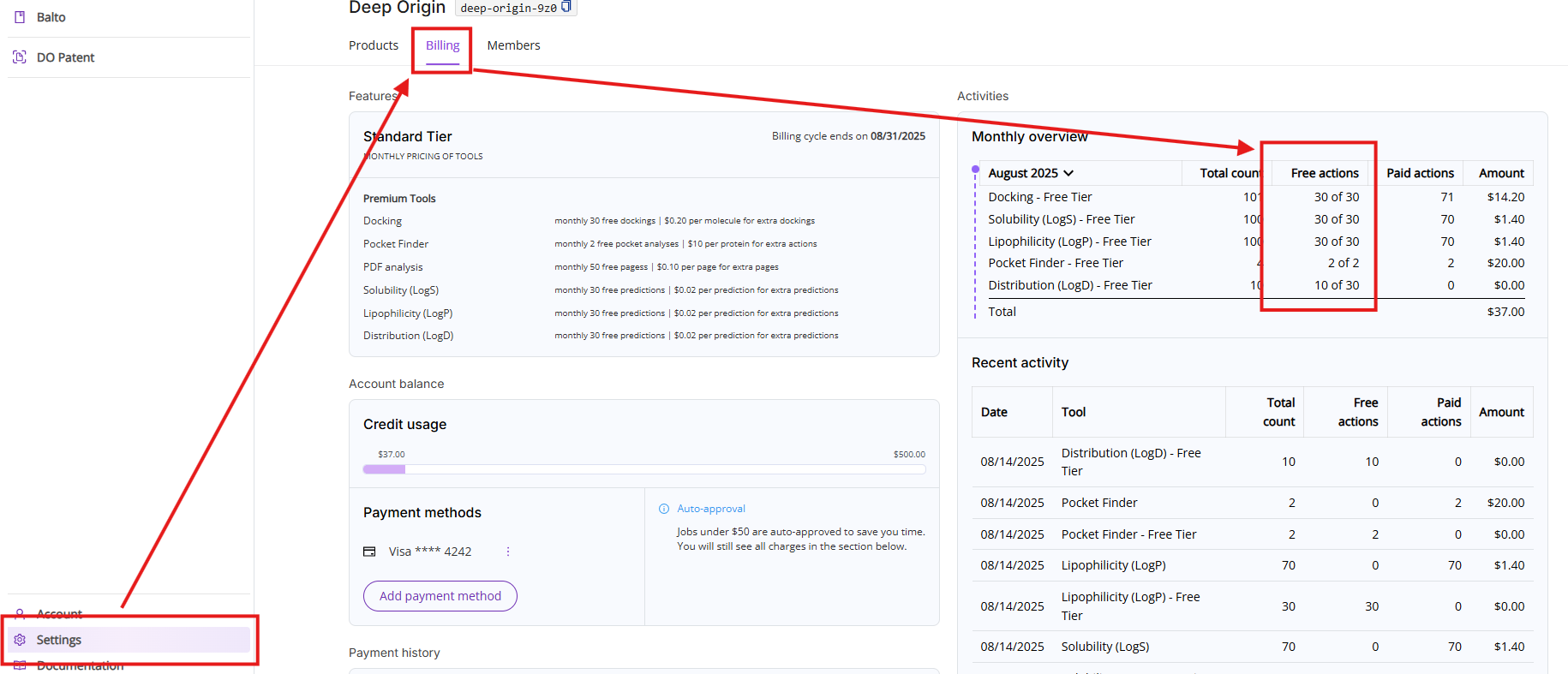
Pricing Tiers
DO Patent has two pricing tiers:
- Standard pricing tier
- Academic pricing tier
You will automatically get an academic tier if you sign up with your .edu account.
Paid Pages
DO Patent has both monthly free pages and an associated cost per page when the free page limit is exceeded. DO Patent charges and free pages appear in the billing view as “PDF analysis”. Free pages are consumed first. Pages exceeding the free limit will be charged according to you Pricing Tier:
- Standard Tier: $0.10 per page
- Academic Tier: $0.06 per page
If you would like to adjust your pricing tier or discuss additional pricing options, then please contact support at support@deeporigin.com.
You can always review your aggregated page count as well as specific actions breakdown by clicking on Settings and then on the Billing tab.
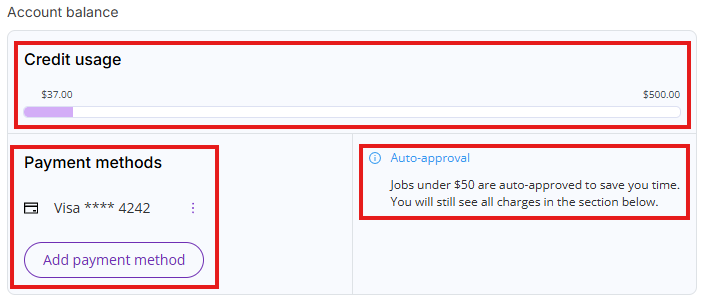
Auto-approval
DO Patent has a default auto-approve threshold set at $50.
DO Patent will automatically execute any actions that will cost less than $50 and will ask for permission to proceed if the job will cost more than $50.
Billing cycle
The credit card on file will be charged after the end of the month for any paid tool actions performed that month.
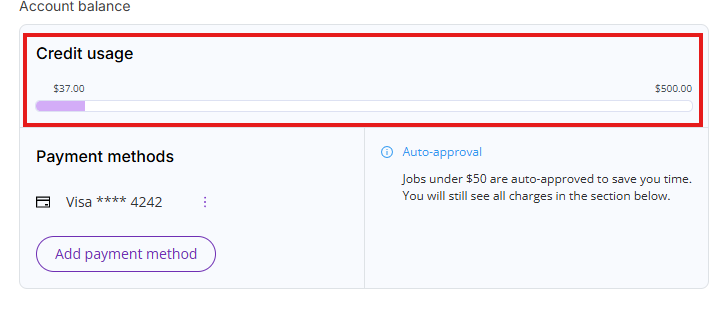
Credit usage
You will automatically receive a $500 credit limit when you enter a payment method. This credit limit allows you to process large PDF documents in excess of the monthly free actions allowance.
The mechanics of this credit limit are similar to a credit card limit. Credit usage accumulates all your unpaid charges for the current month (billing cycle) and unpaid charges for the previous month (billing cycle). Once the bill for the previous billing cycle is paid, your credit usage will be lowered by the paid bill amount. Once you have hit your credit limit, you will not be able to perform additional paid actions without contacting our support team at support@deeporigin.com.
You can always access your credit usage limit by clicking on Settings and then on the Billing tab.
Payments via Purchase Order
Please, contact our customer support team for this request at support@deeporigin.com
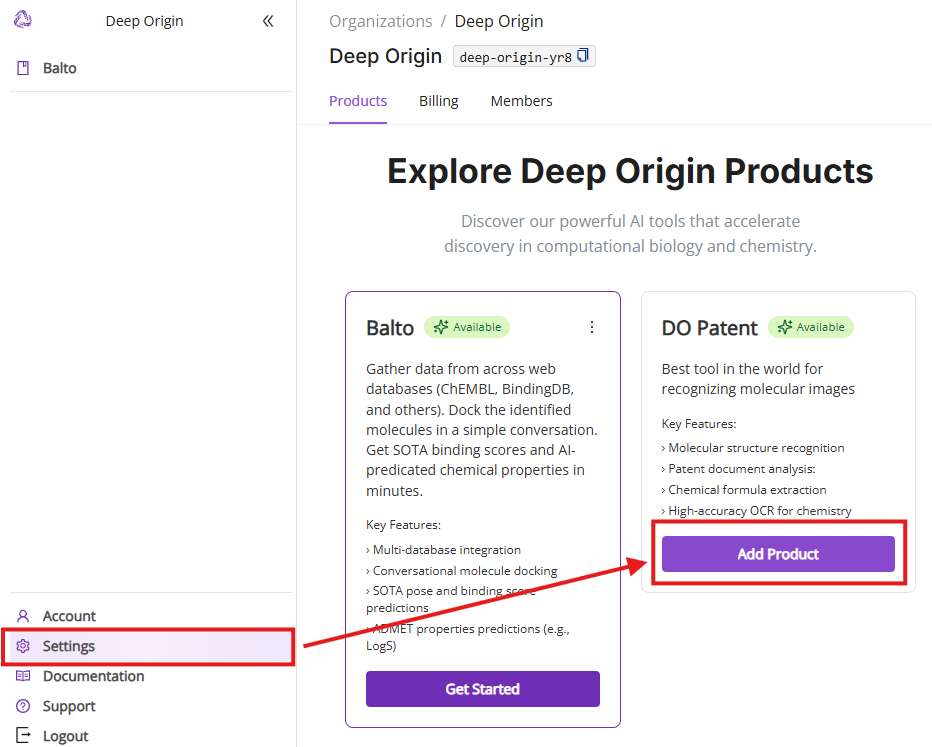
6. Settings Menu
6.1 Products
Clicking “Settings” in the left navigation panel takes you to the Products tab. You can see all available Deep Origin products and add products to your product list. Subscription to additional products is FREE.
6.2 Billing tab
The Billing tab shows details about your current subscription, available tools, current charges and tool usage. The view consists of five sections:
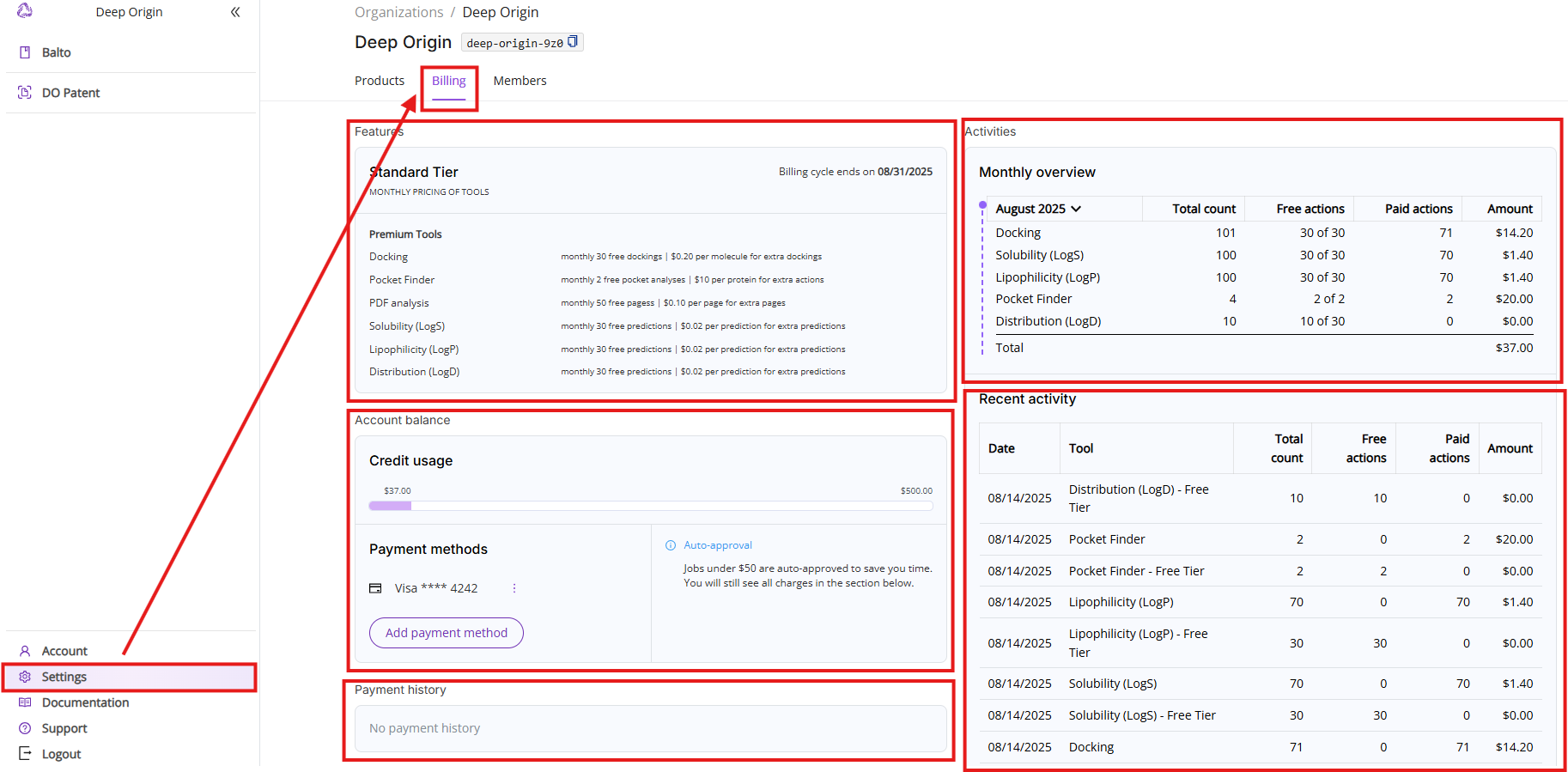
- Features: Shows your current pricing tier, billing cycle and lists available paid tools with free actions limit and pricing.
- Account balance: Shows current credit limit usage, current payment method (if one is set up) and auto-approval threshold (if payment method is set up).
- Payment history: A table displaying past invoices for paid tool usage.
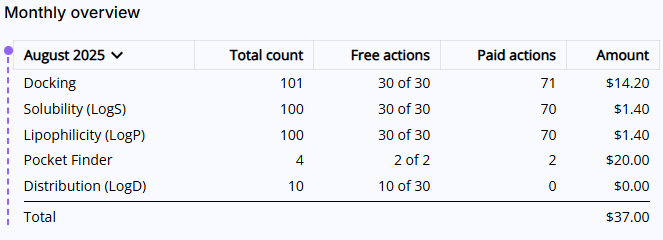
- Monthly overview: Lists all actions executed during the current month. The view is broken down into five columns:
- Period and tool names: You can select a different period by clicking on the month. Note, if a particular tool was not used during this month, it will not show up on this list.
- Total count: Shows total count of free and paid actions.
- Free actions: Shows total count of used free actions and available limit of free actions in the format of XX used of YY available.
- Paid actions: Shows total count of actions above the free actions limit.
- Amount: Shows charges calculated by multiplying your paid actions count and the price of action.
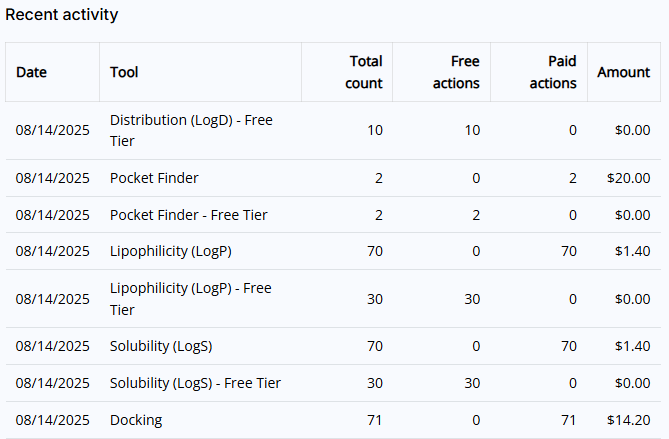
- Recent activities: Shows you a list of the last 10 executed Premium tools actions.
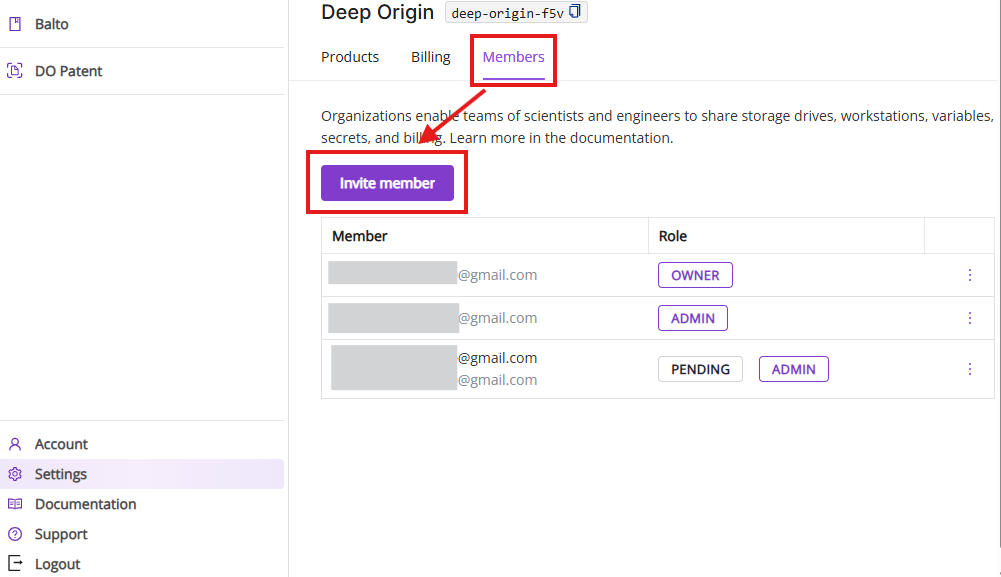
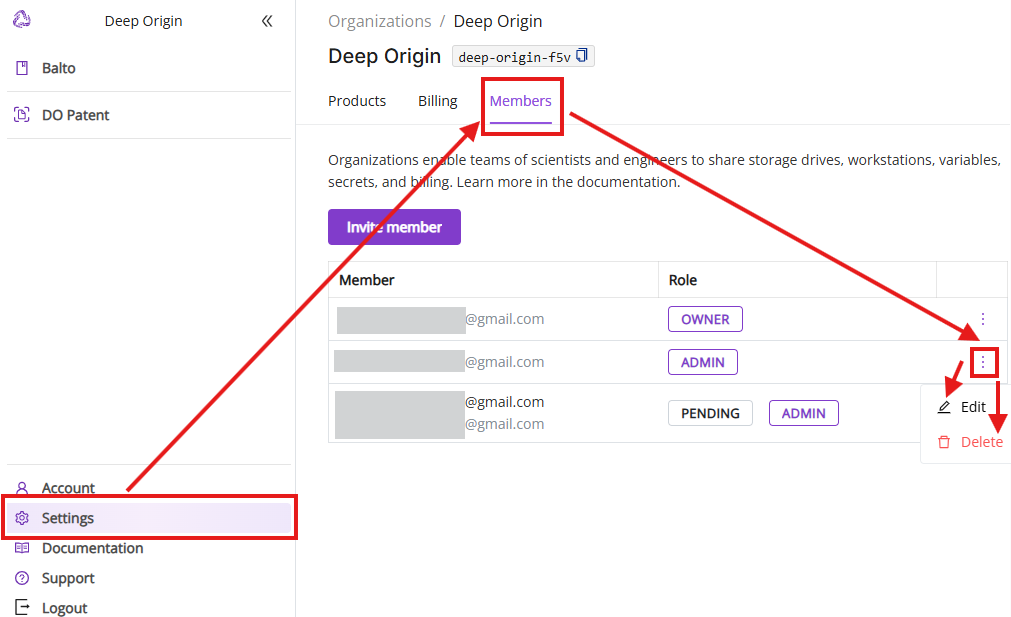
6.3 Members tab
The Members tab shows current members in your Deep Origin Organization, enables inviting and deleting new members and changing their roles.
- Members list: Current members of your organization and their roles (e.g., “Owner”, “Admin”, “Pending”)
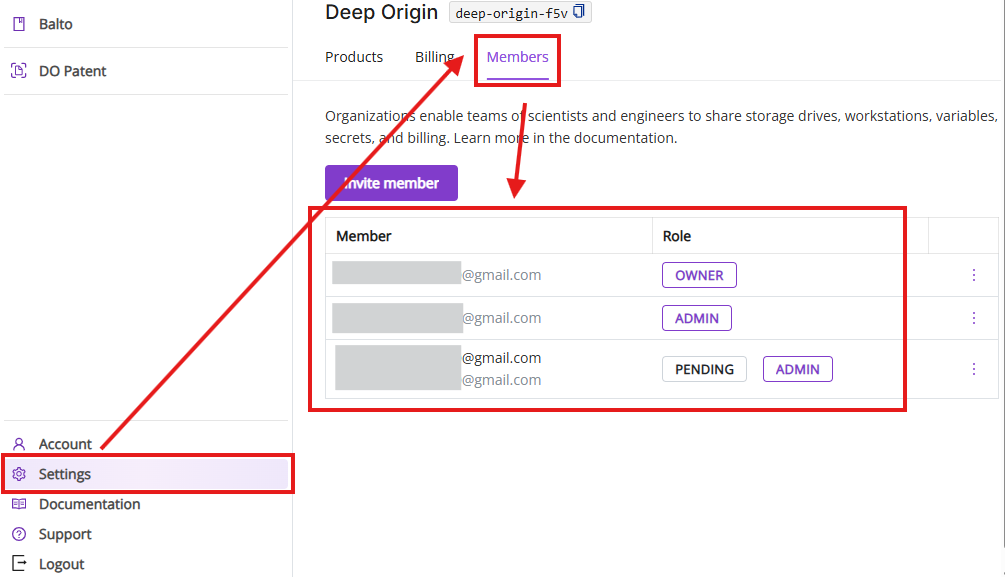
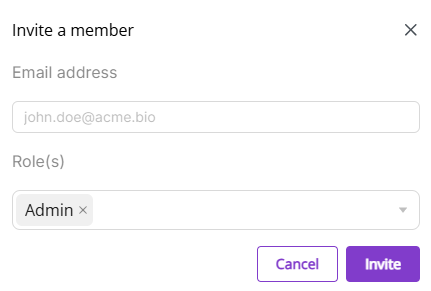
Invite member: Enter email address to invite new members in a pop-up window.
- Edit and delete member: Edit member’s roles and delete existing members from your Deep Origin Organization.
7. Support
For additional guidance, contact us through our support team for assistance at support@deeporigin.com