Downloading files from a workstation
There are several ways to download files from a workstation onto your local computer.
Downloading over SSH (recommended for large files or directories)
SSH is the best way to download large files from your workstation to your local computer. We recommend using rsync, a free tool for copying files. rsync can be installed with the system package manager on Linux/WSL or with Homebrew on Mac. scp is a similar tool without as many features, and comes pre-installed on most Linux distributions and Mac.
- rsync (over SSH)
- WinSCP (over SSH, for Windows users)
- scp (over SSH)
- VS Code server
- JupyterLab
- RStudio Server
We recommend you first complete the advanced SSH configuration to reduce the verbosity in these commands.
Download a single file from a workstation
Run the command below to copy the single file /home/bench-user/sample_1.fastq.gz on your workstation to the fastq directory on your local machine (which must exist already), replacing the following variables:
org-id: Replace with the ID of your organization, such asacme-bio.workstation-id: Replace with the ID of your workstation, such asexceptional-panda-g1i.blueprint-id: Replace with the ID of the blueprint, such aspython.compute-cluster-id: Replace with the ID of your compute cluster, such asus-west-2.aws.ssh-private-key-file: Replace with the path to your private SSH key, such as~/.ssh/id_rsa.
- rsync
- scp
- rsync (with advanced SSH configuration)
- scp (with advanced SSH configuration)
rsync -P -e \
'ssh -i {ssh-private-key-file} -o "ProxyCommand openssl s_client -quiet -connect {compute-cluster-id}.bench.deeporigin.io:2222 -servername {workstation-id}-{blueprint-id}.org-{org-id}"' \
bench-user@{workstation-id}:/home/bench-user/sample_1.fastq.gz \
fastq
scp -o "IdentityFile {ssh-private-key-file}" \
-o "ProxyCommand openssl s_client -quiet -connect {compute-cluster-id}.bench.deeporigin.io:2222 -servername {workstation-id}-{blueprint-id}.org-{org-id}" \
-q \
bench-user@{workstation-id}:/home/bench-user/sample_1.fastq.gz \
fastq
rsync -P {workstation-id}-{blueprint-id}.org-{org-id}.{compute-cluster-id}.bench.deeporigin.io:/home/bench-user/sample_1.fastq.gz fastq/
scp -q {workstation-id}-{blueprint-id}.org-{org-id}.{compute-cluster-id}.bench.deeporigin.io:/home/bench-user/sample_1.fastq.gz fastq
Download a directory from a workstation
Run the command below to copy the directory /home/bench-user/fastq on the workstation to the data directory on the local machine, replacing the following variables:
org-id: Replace with the ID of your organization, such asacme-bio.workstation-id: Replace with the ID of your workstation, such asexceptional-panda-g1i.blueprint-id: Replace with the ID of the blueprint, such aspython.compute-cluster-id: Replace with the ID of your compute cluster, such asus-west-2.aws.ssh-private-key-file: Replace with the path to your private SSH key, such as~/.ssh/id_rsa.
- rsync
- scp
- rsync (with advanced SSH configuration)
- scp (with advanced SSH configuration)
rsync -rP -e \
'ssh -i {ssh-private-key-file} -o "ProxyCommand openssl s_client -quiet -connect {compute-cluster-id}.bench.deeporigin.io:2222 -servername {workstation-id}-{blueprint-id}.org-{org-id}"' \
bench-user@{workstation-id}:/home/bench-user/fastq \
data
It's important to leave the trailing slash off of the remote path fastq, otherwise rsync will copy the contents of fastq, rather than the whole directory.
scp -r \
-o "IdentityFile {ssh-private-key-file}" \
-o "ProxyCommand openssl s_client -quiet -connect {compute-cluster-id}.bench.deeporigin.io:2222 -servername {workstation-id}-{blueprint-id}.org-{org-id}" \
-q \
bench-user@{workstation-id}:/home/bench-user/fastq \
data
rsync -rP {workstation-id}-{blueprint-id}.org-{org-id}.{compute-cluster-id}.bench.deeporigin.io:/home/bench-user/fastq data
It's important to leave the trailing slash off of the remote path fastq, otherwise rsync will copy the contents of fastq, rather than the whole directory.
scp -r -q {workstation-id}-{blueprint-id}.org-{org-id}.{compute-cluster-id}.bench.deeporigin.io:/home/bench-user/fastq data
Helpful rsync options
A few particularly helpful rsync flags are listed below. Use man rsync to get a full explanation of all the options.
--update, -u- This forces rsync to skip any files which exist on the destination and have a modified time that is newer than the source file. (If an existing destination file has a modification time equal to the source file's, it will be updated if the sizes are different.)
-P- The -P option is equivalent to
--partial --progress. Its purpose is to make it much easier to specify these two options for a long transfer that may be interrupted.
- The -P option is equivalent to
--partial- By default, rsync will delete any partially transferred file if the transfer is interrupted. In some circumstances it is more desirable to keep partially transferred files. Using the
--partialoption tells rsync to keep the partial file which should make a subsequent transfer of the rest of the file much faster.
- By default, rsync will delete any partially transferred file if the transfer is interrupted. In some circumstances it is more desirable to keep partially transferred files. Using the
--progress- This option tells rsync to print information showing the progress of the transfer.
--recursive, -r- This tells rsync to copy directories recursively.
--archive, -a- It is a quick way of saying you want recursion and want to preserve almost everything.
--dry-run, -n- This makes rsync perform a trial run that doesn't make any changes (and produces mostly the same output as a real run). It is most commonly used in combination with the
--verbose (-v)and/or--itemize-changes (-i)options to see what an rsync command is going to do before one actually runs it.
- This makes rsync perform a trial run that doesn't make any changes (and produces mostly the same output as a real run). It is most commonly used in combination with the
Downloading with WinSCP
WinSCP is a free tool for managing files on remote servers from a Windows computer. Please follow the instructions for uploading to set up a WinSCP connection.
Downloading when connected to VS Code over SSH
- Connect to your workstation with VS Code over SSH.
- Navigate to the location on the workstation with the file to download, either through the "Explorer" tab, or by selecting "File > Open Folder" from the menu.
- In the "Explorer" tab on the left side of the interface, right click the file and select "Download".
- Select a target location on your local computer and click "Download".
- The status bar at the bottom of the VS Code window will indicate the download status.
Downloading over the web
Many of the web endpoints provided for workstations have a way to download single files. These may be slower than SSH, don't offer the ability to download folders, and can't selectively download newer files or resume failed transfers.
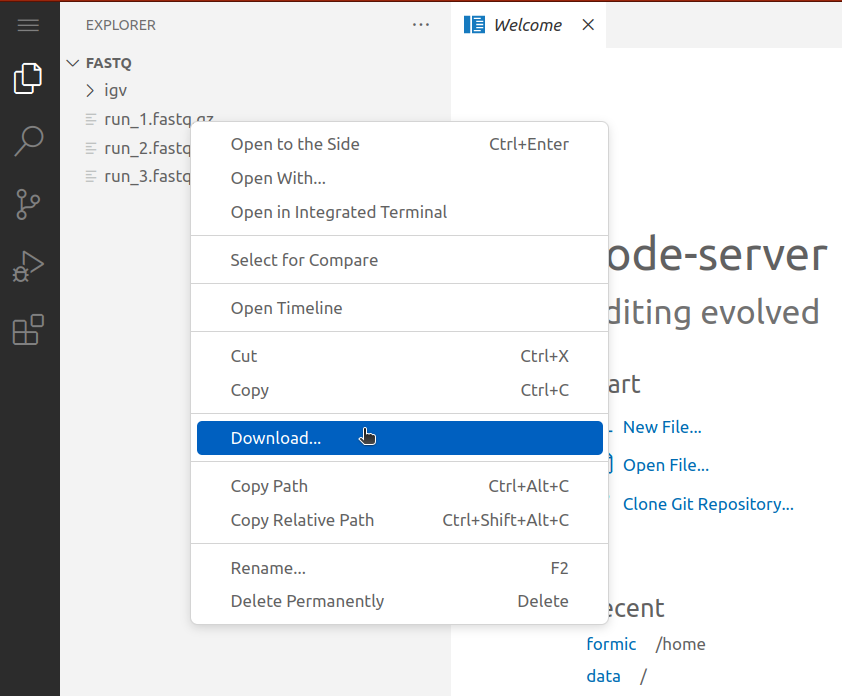
Downloading files in VS Code server
- Open the VS Code server web endpoint from the workstation list.
- Navigate to the location on the workstation with the file to download, either through the "Explorer" tab, or by selecting "File > Open Folder" from the menu.
- In the "Explorer" tab on the left side of the interface, right click the file and select "Download".
- The file will appear in your browser's download queue.
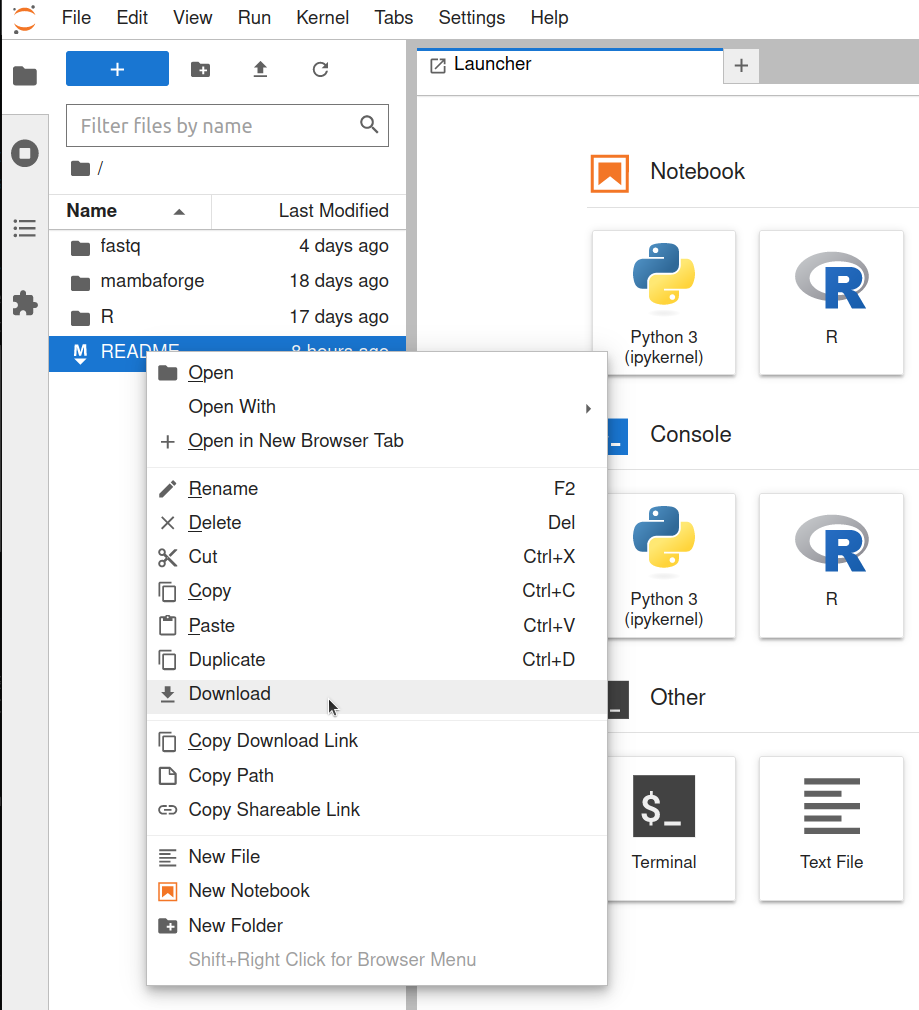
Downloading files in JupyterLab
- Open the JupyterLab web endpoint from the workstation list.
- From the "File Browser" sidebar, locate the file you want to download, and right click it.
- Select "Download" from the menu.
- The file will appear in your browser's download queue.
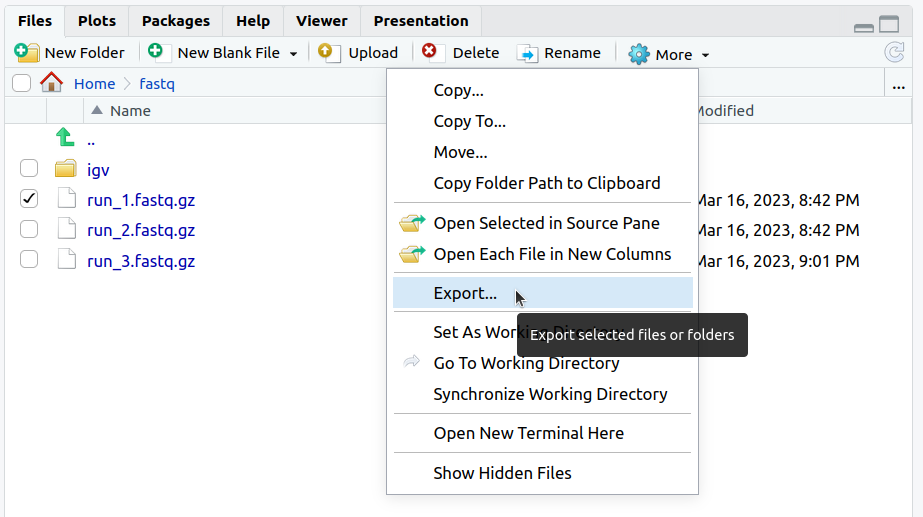
Downloading files in RStudio Server
- Open the RStudio Server web endpoint from the workstation list.
- From the "File" sidebar, locate and select the files and folders you want to download. RStudio will create a zip file if the selection contains more than one file.
- Select More > Export from the toolbar
- Specify the file name for the download and click OK.
- The file will appear in your browser's download queue.